Evaluation of regression models in metabolic physiology: predicting fluxes from isotopic data without knowledge of the pathway
- PMID: 17066125
- PMCID: PMC1622920
- DOI: 10.1007/s11306-006-0018-2
Evaluation of regression models in metabolic physiology: predicting fluxes from isotopic data without knowledge of the pathway
Abstract
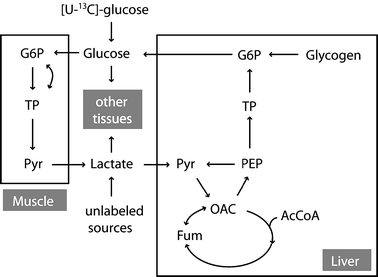
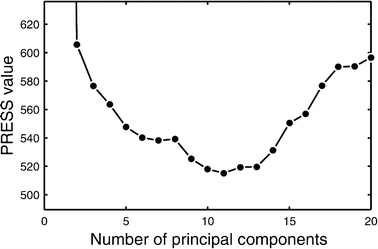
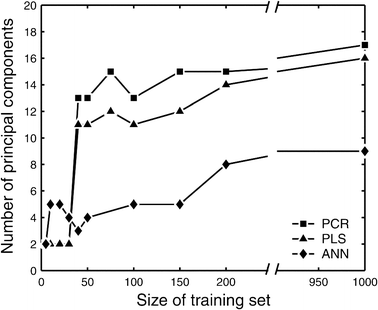
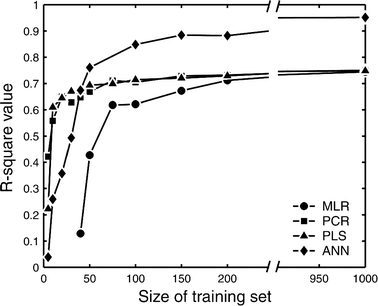
This study explores the ability of regression models, with no knowledge of the underlying physiology, to estimate physiological parameters relevant for metabolism and endocrinology. Four regression models were compared: multiple linear regression (MLR), principal component regression (PCR), partial least-squares regression (PLS) and regression using artificial neural networks (ANN). The pathway of mammalian gluconeogenesis was analyzed using [U-(13)C]glucose as tracer. A set of data was simulated by randomly selecting physiologically appropriate metabolic fluxes for the 9 steps of this pathway as independent variables. The isotope labeling patterns of key intermediates in the pathway were then calculated for each set of fluxes, yielding 29 dependent variables. Two thousand sets were created, allowing independent training and test data. Regression models were asked to predict the nine fluxes, given only the 29 isotopomers. For large training sets (>50) the artificial neural network model was superior, capturing 95% of the variability in the gluconeogenic flux, whereas the three linear models captured only 75%. This reflects the ability of neural networks to capture the inherent non-linearities of the metabolic system. The effect of error in the variables and the addition of random variables to the data set was considered. Model sensitivities were used to find the isotopomers that most influenced the predicted flux values. These studies provide the first test of multivariate regression models for the analysis of isotopomer flux data. They provide insight for metabolomics and the future of isotopic tracers in metabolic research where the underlying physiology is complex or unknown.
Figures
Similar articles
-
Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions.Metab Eng. 2007 Jan;9(1):68-86. doi: 10.1016/j.ymben.2006.09.001. Epub 2006 Sep 17. Metab Eng. 2007. PMID: 17088092 Free PMC article.
-
Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. PMID: 26269925 Free Books & Documents. Review.
-
Predicting blood β-hydroxybutyrate using milk Fourier transform infrared spectrum, milk composition, and producer-reported variables with multiple linear regression, partial least squares regression, and artificial neural network.J Dairy Sci. 2018 May;101(5):4378-4387. doi: 10.3168/jds.2017-14076. Epub 2018 Feb 22. J Dairy Sci. 2018. PMID: 29477523
-
Designer labels for plant metabolism: statistical design of isotope labeling experiments for improved quantification of flux in complex plant metabolic networks.Mol Biosyst. 2013 Jan 27;9(1):99-112. doi: 10.1039/c2mb25253h. Epub 2012 Nov 1. Mol Biosyst. 2013. PMID: 23114423
-
Probing metabolic pathways with isotopic tracers: insights from mammalian metabolic physiology.Metab Eng. 2004 Jan;6(1):1-5. doi: 10.1016/j.ymben.2003.10.008. Metab Eng. 2004. PMID: 14734250 Review.
Cited by
-
A unified framework for estimating parameters of kinetic biological models.BMC Bioinformatics. 2015 Mar 27;16:104. doi: 10.1186/s12859-015-0500-9. BMC Bioinformatics. 2015. PMID: 25886743 Free PMC article.
-
Carbon-flux distribution within Streptomyces coelicolor metabolism: a comparison between the actinorhodin-producing strain M145 and its non-producing derivative M1146.PLoS One. 2013 Dec 23;8(12):e84151. doi: 10.1371/journal.pone.0084151. eCollection 2013. PLoS One. 2013. PMID: 24376790 Free PMC article.
-
Predicting metabolic fluxes from omics data via machine learning: Moving from knowledge-driven towards data-driven approaches.Comput Struct Biotechnol J. 2023 Oct 5;21:4960-4973. doi: 10.1016/j.csbj.2023.10.002. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37876626 Free PMC article.
-
Metabolomics-driven nutraceutical evaluation of diverse green tea cultivars.PLoS One. 2011;6(8):e23426. doi: 10.1371/journal.pone.0023426. Epub 2011 Aug 10. PLoS One. 2011. PMID: 21853132 Free PMC article.
-
Non-linearity of Metabolic Pathways Critically Influences the Choice of Machine Learning Model.Front Artif Intell. 2022 Jun 10;5:744755. doi: 10.3389/frai.2022.744755. eCollection 2022. Front Artif Intell. 2022. PMID: 35757298 Free PMC article.
References
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'PubMed', 'value': '8203653', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/8203653/'}]}
- Benigni R., Giuliani A. (1994) Quantitative modeling and biology: the multivariate approach. Am. J. Physiol 266:R1697–R1704 - PubMed
-
- Bishop, C.M. (1996). Neural Networks for Pattern Recognition. Oxford University Press.
-
- None
- Causton D.R. (1987). A Biologist’s Advanced Mathematics. London, Allen & Unwin
-
- None
- Chatfield C., Collins A.J. (1981). Introduction to Multivariate Analysis. London, Chapman & Hall
-
- None
- Dillon W.R., Goldstein M. (1984) Multivariate Analysis Methods and Applications. Wiley, New York
Grants and funding
LinkOut - more resources
Full Text Sources
