454 sequencing put to the test using the complex genome of barley
- PMID: 17067373
- PMCID: PMC1633745
- DOI: 10.1186/1471-2164-7-275
454 sequencing put to the test using the complex genome of barley
Abstract
Background: During the past decade, Sanger sequencing has been used to completely sequence hundreds of microbial and a few higher eukaryote genomes. In recent years, a number of alternative technologies became available, among them adaptations of the pyrosequencing procedure (i.e. "454 sequencing"), promising an approximately 100-fold increase in throughput over Sanger technology--an advancement which is needed to make large and complex genomes more amenable to full genome sequencing at affordable costs. Although several studies have demonstrated its potential usefulness for sequencing small and compact microbial genomes, it was unclear how the new technology would perform in large and highly repetitive genomes such as those of wheat or barley.
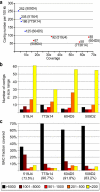
Results: To study its performance in complex genomes, we used 454 technology to sequence four barley Bacterial Artificial Chromosome (BAC) clones and compared the results to those from ABI-Sanger sequencing. All gene containing regions were covered efficiently and at high quality with 454 sequencing whereas repetitive sequences were more problematic with 454 sequencing than with ABI-Sanger sequencing. 454 sequencing provided a much more even coverage of the BAC clones than ABI-Sanger sequencing, resulting in almost complete assembly of all genic sequences even at only 9 to 10-fold coverage. To obtain highly advanced working draft sequences for the BACs, we developed a strategy to assemble large parts of the BAC sequences by combining comparative genomics, detailed repeat analysis and use of low-quality reads from 454 sequencing. Additionally, we describe an approach of including small numbers of ABI-Sanger sequences to produce hybrid assemblies to partly compensate the short read length of 454 sequences.
Conclusion: Our data indicate that 454 pyrosequencing allows rapid and cost-effective sequencing of the gene-containing portions of large and complex genomes and that its combination with ABI-Sanger sequencing and targeted sequence analysis can result in large regions of high-quality finished genomic sequences.
Figures


References
-
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer MLI, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

