Global analysis of mRNA stability in the archaeon Sulfolobus
- PMID: 17067383
- PMCID: PMC1794556
- DOI: 10.1186/gb-2006-7-10-r99
Global analysis of mRNA stability in the archaeon Sulfolobus
Abstract
Background: Transcript half-lives differ between organisms, and between groups of genes within the same organism. The mechanisms underlying these differences are not clear, nor are the biochemical properties that determine the stability of a transcript. To address these issues, genome-wide mRNA decay studies have been conducted in eukaryotes and bacteria. In contrast, relatively little is known about RNA stability in the third domain of life, Archaea. Here, we present a microarray-based analysis of mRNA half-lives in the hyperthermophilic crenarchaea Sulfolobus solfataricus and Sulfolobus acidocaldarius, constituting the first genome-wide study of RNA decay in archaea.
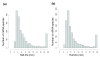
Results: The two transcriptomes displayed similar half-life distributions, with medians of about five minutes. Growth-related genes, such as those involved in transcription, translation and energy production, were over-represented among unstable transcripts, whereas uncharacterized genes were over-represented among the most stable. Half-life was negatively correlated with transcript abundance and, unlike the situation in other organisms, also negatively correlated with transcript length.
Conclusion: The mRNA half-life distribution of Sulfolobus species is similar to those of much faster growing bacteria, contrasting with the earlier observation that median mRNA half-life is proportional to the minimal length of the cell cycle. Instead, short half-lives may be a general feature of prokaryotic transcriptomes, possibly related to the absence of a nucleus and/or more limited post-transcriptional regulatory mechanisms. The pattern of growth-related transcripts being among the least stable in Sulfolobus may also indicate that the short half-lives reflect a necessity to rapidly reprogram gene expression upon sudden changes in environmental conditions.
Figures
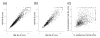
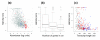
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

