Babel's tower revisited: a universal resource for cross-referencing across annotation databases
- PMID: 17068090
- PMCID: PMC2435247
- DOI: 10.1093/bioinformatics/btl372
Babel's tower revisited: a universal resource for cross-referencing across annotation databases
Abstract
Motivation: Annotation databases are widely used as public repositories of biological knowledge. However, most of these resources have been developed by independent groups which used different designs and different identifiers for the same biological entities. As we show in this article, incoherent name spaces between various databases represent a serious impediment to using the existing annotations at their full potential. Navigating between various such name spaces by mapping IDs from one database to another is a very important issue which is not properly addressed at the moment.
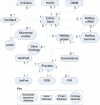
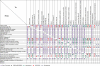
Results: We have developed a web-based resource, Onto-Translate (OT), which effectively addresses this problem. OT is able to map onto each other different types of biological entities from the following annotation databases: Swiss-Prot, TrEMBL, NREF, PIR, Gene Ontology, KEGG, Entrez Gene, GenBank, GenPept, IMAGE, RefSeq, UniGene, OMIM, PDB, Eukaryotic Promoter Database, HUGO Gene Nomenclature Committee and NetAffx. Currently, OT is able to perform 462 types of mappings between 29 different types of IDs from 17 databases concerning 53 organisms. Among these, over 300 types of translations and 15 types of IDs are not currently supported by any other tool or resource. On average, OT is able to correctly map between 96 and 99% of the biological entities provided as input. In terms of speed, sets of approximately 20 000 IDs can be translated in <30 s, in most cases.
Availability: OT is a part of Onto-Tools, which is freely available at http://vortex.cs.wayne.edu/Projects.html
Figures
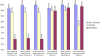

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

