Legionella pneumophila glucosyltransferase inhibits host elongation factor 1A
- PMID: 17068130
- PMCID: PMC1636560
- DOI: 10.1073/pnas.0601562103
Legionella pneumophila glucosyltransferase inhibits host elongation factor 1A
Abstract
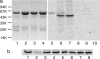
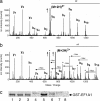
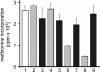
Legionella pneumophila, the causal agent of Legionnaires' disease, is an intracellular parasite and invades and proliferates within different eukaryotic cells, including human alveolar macrophages. After several 100-fold multiplication within host cells, the pathogens are released for new invasion by induction of apoptosis or necrosis. Here we report that L. pneumophila produces a glucosyltransferase, which selectively modifies an approximately 50-kDa mammalian protein by using UDP-glucose as a cosubstrate. MS analysis identified the protein substrate as the mammalian elongation factor (EF)1A. Legionella glucosyltransferase modifies its eukaryotic protein substrate at serine-53, which is located in the GTPase domain of the EF. Glucosylation of EF1A results in inhibition of eukaryotic protein synthesis and death of target cells. Our findings show a mode of inhibition of protein synthesis by microbial pathogens and offer a perspective for understanding of the host-pathogen interaction of L. pneumophila.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

