Structure-guided mutational analysis of T4 RNA ligase 1
- PMID: 17068206
- PMCID: PMC1664725
- DOI: 10.1261/rna.271706
Structure-guided mutational analysis of T4 RNA ligase 1
Abstract
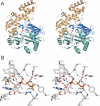
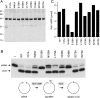
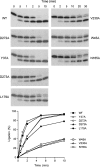
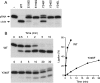
T4 RNA ligase 1 (Rnl1) is a tRNA repair enzyme that circumvents an RNA-damaging host antiviral response. Whereas the three-step reaction scheme of Rnl1 is well established, the structural basis for catalysis has only recently been appreciated as mutational and crystallographic approaches have converged. Here we performed a structure-guided alanine scan of nine conserved residues, including side chains that either contact the ATP substrate via adenine (Leu179, Val230), the 2'-OH (Glu159), or the gamma phosphate (Tyr37) or coordinate divalent metal ions at the ATP alpha phosphate (Glu159, Tyr246) or beta phosphate (Asp272, Asp273). We thereby identified Glu159 and Tyr246 as essential for RNA sealing activity in vitro and for tRNA repair in vivo. Structure-activity relationships at Glu159 and Tyr246 were clarified by conservative substitutions. Eliminating the phosphate-binding Tyr37, and the magnesium-binding Asp272 and Asp273 side chains had little impact on sealing activity in vitro or in vivo, signifying that not all atomic interactions in the active site are critical for function. Analysis of mutational effects on individual steps of the ligation pathway underscored how different functional groups come into play during the ligase-adenylylation reaction versus the subsequent steps of RNA-adenylylation and phosphodiester formation. Moreover, the requirements for sealing exogenous preformed RNA-adenylate are more stringent than are those for sealing the RNA-adenylate intermediate formed in situ during ligation of a 5'-PO4 RNA.
Figures


References
-
- Blondal, T., Thorisdottir, A., Unnsteinsdottir, U., Hjorleifsdottir, S., Aevarsson, A., Ernstsson, S., Fridjonsson, O.H., Skirnisdottir, S., Wheat, J.O., Hermannsdottir, A.G., et al. Isolation and characterization of a thermostable RNA ligase 1 from a Thermus scotoductus bacteriophage TS2126 with good single-stranded DNA ligation properties. Nucleic Acids Res. 2005;33:135–142. - PMC - PubMed
-
- Cranston, J.W., Silber, R., Malathi, V.G., Hurwitz, J. Studies on ribonucleic acid ligase: Characterization of an adenosine triphosphate-inorganic pyrophosphate exchange reaction and demonstration of an enzyme-adenylate complex with T4 bacteriophage-induced enzyme. J. Biol. Chem. 1974;249:7447–7456. - PubMed
-
- Deng, J., Schnaufer, A., Salavati, R., Stuart, K.D., Hol, W.G.J. High resolution crystal structure of a key editosome enzyme from Trypanosoma brucei: RNA editing ligase 1. J. Mol. Biol. 2004;343:601–613. - PubMed
-
- El Omari, K., Ren, J., Bird, L.E., Bona, M.K., Klarmann, G., LeGrice, S.F.J., Stammers, D.K. Molecular architecture and ligand recognition determinants for T4 RNA ligase. J. Biol. Chem. 2006;281:1573–1579. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
