Screening for plant transporter function by expressing a normalized Arabidopsis full-length cDNA library in Xenopus oocytes
- PMID: 17069646
- PMCID: PMC1637106
- DOI: 10.1186/1746-4811-2-17
Screening for plant transporter function by expressing a normalized Arabidopsis full-length cDNA library in Xenopus oocytes
Abstract
Background: We have developed a functional genomics approach based on expression cloning in Xenopus oocytes to identify plant transporter function. We utilized the full-length cDNA databases to generate a normalized library consisting of 239 full-length Arabidopsis thaliana transporter cDNAs. The genes were arranged into a 96-well format and optimized for expression in Xenopus oocytes by cloning each coding sequence into a Xenopus expression vector.
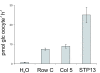
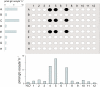
Results: Injection of 96 in vitro transcribed cRNAs from the library in pools of columns and rows into oocytes and subsequent screening for glucose uptake activity identified three glucose transporters. One of these, AtSTP13, had not previously been experimentally characterized.
Conclusion: Expression of the library in Xenopus oocytes, combined with uptake assays, has great potential in assignment of plant transporter function and for identifying membrane transporters for the many plant metabolites where a transporter has not yet been identified.
Figures

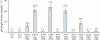
References
-
- The Institute for Genomic Research (TIGR) http://www.tigr.org/tdb/e2k1/ath1/ath1.shtml
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

