Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior
- PMID: 17071647
- PMCID: PMC1676044
- DOI: 10.1104/pp.106.088534
Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior
Abstract
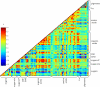
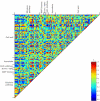
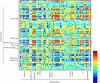
Tomato (Solanum lycopersicum) is a well-studied model of fleshy fruit development and ripening. Tomato fruit development is well understood from a hormonal-regulatory perspective, and developmental changes in pigment and cell wall metabolism are also well characterized. However, more general aspects of metabolic change during fruit development have not been studied despite the importance of metabolism in the context of final composition of the ripe fruit. In this study, we quantified the abundance of a broad range of metabolites by gas chromatography-mass spectrometry, analyzed a number of the principal metabolic fluxes, and in parallel analyzed transcriptomic changes during tomato fruit development. Metabolic profiling revealed pronounced shifts in the abundance of metabolites of both primary and secondary metabolism during development. The metabolite changes were reflected in the flux analysis that revealed a general decrease in metabolic activity during ripening. However, there were several distinct patterns of metabolite profile, and statistical analysis demonstrated that metabolites in the same (or closely related) pathways changed in abundance in a coordinated manner, indicating a tight regulation of metabolic activity. The metabolite data alone allowed investigations of likely routes through the metabolic network, and, as an example, we analyze the operational feasibility of different pathways of ascorbate synthesis. When combined with the transcriptomic data, several aspects of the regulation of metabolism during fruit ripening were revealed. First, it was apparent that transcript abundance was less strictly coordinated by functional group than metabolite abundance, suggesting that posttranslational mechanisms dominate metabolic regulation. Nevertheless, there were some correlations between specific transcripts and metabolites, and several novel associations were identified that could provide potential targets for manipulation of fruit compositional traits. Finally, there was a strong relationship between ripening-associated transcripts and specific metabolite groups, such as TCA-cycle organic acids and sugar phosphates, underlining the importance of the respective metabolic pathways during fruit development.
Figures



References
-
- Adams-Phillips L, Barry C, Giovannoni J (2004) Signal transduction systems regulating fruit development. Trends Plant Sci 9: 331–338 - PubMed
-
- Agius F, Gonzalez-Lemothe R, Caballero JL, Munoz-Blanco J, Botella MA, Valpuesta V (2003) Engineering increased vitamin C levels in plants by overexpression of a D-galacturonic acid reductase. Nat Biotechnol 21: 177–181 - PubMed
-
- Alba R, Fei ZJ, Payton P, Liu Y, Moore SL, Debbie P, Cohn J, D'Ascenzo M, Gordon JS, Rose JK, et al (2004) ESTs, cDNA microarrays, and gene expression profiling: tools for dissecting plant physiology and development. Plant J 39: 697–714 - PubMed
-
- Albersheim P, Nevins DJ, English PD, Karr A (1967) A method for the analysis of sugars in plant cell wall polysaccharides by gas-liquid chromatography. Carbohydr Res 5: 340–345
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

