Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns
- PMID: 17071648
- PMCID: PMC1676066
- DOI: 10.1104/pp.106.088096
Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns
Abstract
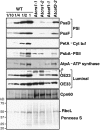
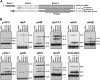
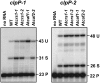
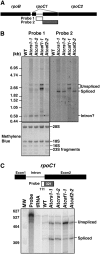
Chloroplast genomes in plants and green algae contain numerous group II introns, large ribozymes that splice via the same chemical steps as spliceosome-mediated splicing in the nucleus. Most chloroplast group II introns are degenerate, requiring interaction with nucleus-encoded proteins to splice in vivo. Genetic approaches in maize (Zea mays) and Chlamydomonas reinhardtii have elucidated distinct sets of proteins that assemble with chloroplast group II introns and facilitate splicing. Little information is available, however, concerning these processes in Arabidopsis (Arabidopsis thaliana). To determine whether the paucity of data concerning chloroplast splicing factors in Arabidopsis reflects a fundamental difference between protein-facilitated group II splicing in monocot and dicot plants, we examined the mutant phenotypes associated with T-DNA insertions in Arabidopsis genes encoding orthologs of the maize chloroplast splicing factors CRS1, CAF1, and CAF2 (AtCRS1, AtCAF1, and AtCAF2). We show that the splicing functions and intron specificities of these proteins are largely conserved between maize and Arabidopsis, indicating that these proteins were recruited to promote the splicing of plastid group II introns prior to the divergence of monocot and dicot plants. We show further that AtCAF1 promotes the splicing of two group II introns, rpoC1 and clpP-intron 1, that are found in Arabidopsis but not in maize; AtCAF1 is the first splicing factor described for these introns. Finally, we show that a strong AtCAF2 allele conditions an embryo-lethal phenotype, adding to the body of data suggesting that cell viability is more sensitive to the loss of plastid translation in Arabidopsis than in maize.
Figures

References
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Barkan A (1998) Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol 297: 38–57
-
- Barkan A (2004) Intron splicing in plant organelles. In H Daniell, C Chase, eds, Molecular Biology and Biotechnology of Plant Organelles. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 281–308
-
- Cahoon A, Cunningham K, Stern D (2003) The plastid clpP gene may not be essential for plant cell viability. Plant Cell Physiol 44: 93–95 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

