The yefM-yoeB toxin-antitoxin systems of Escherichia coli and Streptococcus pneumoniae: functional and structural correlation
- PMID: 17071753
- PMCID: PMC1797350
- DOI: 10.1128/JB.01130-06
The yefM-yoeB toxin-antitoxin systems of Escherichia coli and Streptococcus pneumoniae: functional and structural correlation
Abstract
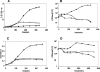
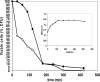
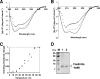
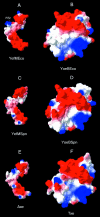
Toxin-antitoxin loci belonging to the yefM-yoeB family are located in the chromosome or in some plasmids of several bacteria. We cloned the yefM-yoeB locus of Streptococcus pneumoniae, and these genes encode bona fide antitoxin (YefM(Spn)) and toxin (YoeB(Spn)) products. We showed that overproduction of YoeB(Spn) is toxic to Escherichia coli cells, leading to severe inhibition of cell growth and to a reduction in cell viability; this toxicity was more pronounced in an E. coli B strain than in two E. coli K-12 strains. The YoeB(Spn)-mediated toxicity could be reversed by the cognate antitoxin, YefM(Spn), but not by overproduction of the E. coli YefM antitoxin. The pneumococcal proteins were purified and were shown to interact with each other both in vitro and in vivo. Far-UV circular dichroism analyses indicated that the pneumococcal antitoxin was partially, but not totally, unfolded and was different than its E. coli counterpart. Molecular modeling showed that the toxins belonging to the family were homologous, whereas the antitoxins appeared to be specifically designed for each bacterial locus; thus, the toxin-antitoxin interactions were adapted to the different bacterial environmental conditions. Both structural features, folding and the molecular modeled structure, could explain the lack of cross-complementation between the pneumococcal and E. coli antitoxins.
Figures

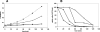

References
-
- Andrade, M. A., P. Chacón, J. J. Merolo, and F. Morán. 1993. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 6:443-454. - PubMed
-
- Baquero, F. 1996. Epidemiology and management of penicillin-resistant pneumococci. Curr. Opin. Infect. Dis. 9:372-379.
-
- Bravo, A., G. de Torrontegui, and R. Díaz. 1987. Identification of components of a new stability system of plasmid R1, Par D, that is close to the origin of replication of this plasmid. Mol. Gen. Genet. 210:101-110. - PubMed
-
- Cherny, I., and E. Gazit. 2004. The YefM antitoxin defines a family of natively unfolded proteins: implications as a novel antibacterial target. J. Biol. Chem. 279:8252-8261. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

