Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR
- PMID: 17071756
- PMCID: PMC1797413
- DOI: 10.1128/JB.00981-06
Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR
Abstract
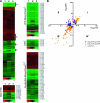
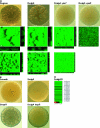
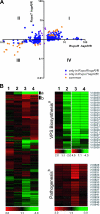
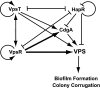
Vibrio cholerae undergoes phenotypic variation that generates two morphologically different variants, termed smooth and rugose. The transcriptional profiles of the two variants differ greatly, and many of the differentially regulated genes are controlled by a complex regulatory circuitry that includes the transcriptional regulators VpsR, VpsT, and HapR. In this study, we identified the VpsT regulon and compared the VpsT and VpsR regulons to elucidate the contribution of each positive regulator to the rugose variant transcriptional profile and associated phenotypes. We have found that although the VpsT and VpsR regulons are very similar, the magnitude of the gene regulation accomplished by each regulator is different. We also determined that cdgA, which encodes a GGDEF domain protein, is partially responsible for the altered vps gene expression between the vpsT and vpsR mutants. Analysis of epistatic relationships among hapR, vpsT, and vpsR with respect to a whole-genome expression profile, colony morphology, and biofilm formation revealed that vpsR is epistatic to hapR and vpsT. Expression of virulence genes was increased in a vpsR hapR double mutant relative to a hapR mutant, suggesting that VpsR negatively regulates virulence gene expression in the hapR mutant. These results show that a complex regulatory interplay among VpsT, VpsR, HapR, and GGDEF/EAL family proteins controls transcription of the genes required for Vibrio polysaccharide and virulence factor production in V. cholerae.
Figures


References
-
- Ali, A., J. A. Johnson, A. A. Franco, D. J. Metzger, T. D. Connell, J. G. Morris, Jr., and S. Sozhamannan. 2000. Mutations in the extracellular protein secretion pathway genes (eps) interfere with rugose polysaccharide production in and motility of Vibrio cholerae. Infect. Immun. 68:1967-1974. - PMC - PubMed
-
- Bao, Y., D. P. Lies, H. Fu, and G. P. Roberts. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167-168. - PubMed
-
- Carroll, P. A., K. T. Tashima, M. B. Rogers, V. J. DiRita, and S. B. Calderwood. 1997. Phase variation in tcpH modulates expression of the ToxR regulon in Vibrio cholerae. Mol. Microbiol. 25:1099-1111. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

