Manipulation of the nuclear factor-kappaB pathway and the innate immune response by viruses
- PMID: 17072332
- PMCID: PMC7100320
- DOI: 10.1038/sj.onc.1209941
Manipulation of the nuclear factor-kappaB pathway and the innate immune response by viruses
Abstract
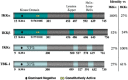
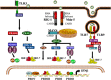
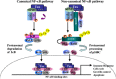
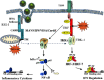
Viral and microbial constituents contain specific motifs or pathogen-associated molecular patterns (PAMPs) that are recognized by cell surface- and endosome-associated Toll-like receptors (TLRs). In addition, intracellular viral double-stranded RNA is detected by two recently characterized DExD/H box RNA helicases, RIG-I and Mda-5. Both TLR-dependent and -independent pathways engage the IkappaB kinase (IKK) complex and related kinases TBK-1 and IKKvarepsilon. Activation of the nuclear factor kappaB (NF-kappaB) and interferon regulatory factor (IRF) transcription factor pathways are essential immediate early steps of immune activation; as a result, both pathways represent prime candidates for viral interference. Many viruses have developed strategies to manipulate NF-kappaB signaling through the use of multifunctional viral proteins that target the host innate immune response pathways. This review discusses three rapidly evolving areas of research on viral pathogenesis: the recognition and signaling in response to virus infection through TLR-dependent and -independent mechanisms, the involvement of NF-kappaB in the host innate immune response and the multitude of strategies used by different viruses to short circuit the NF-kappaB pathway.
Figures

References
-
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D . (2000). Cell103: 667–678. - PubMed
-
- Aizaki H, Saito S, Ogino T, Miyajima N, Harada T, Matsuura Y et al. (2000). J Interferon Cytokine Res20: 1111–1120. - PubMed
-
- Akagi T, Ono H, Shinotohno K . (1995). Blood86: 4243–4249. - PubMed
-
- Akira S, Sato S . (2003). Scand J Infect Dis35: 555–562. - PubMed
-
- Akira S, Uematsu S, Takeuchi O . (2006). Cell124: 783–801. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

