Structural analysis of the catalytic core of human telomerase RNA by FRET and molecular modeling
- PMID: 17073451
- PMCID: PMC2196208
- DOI: 10.1021/bi061150a
Structural analysis of the catalytic core of human telomerase RNA by FRET and molecular modeling
Abstract
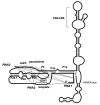
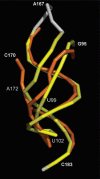
Telomerase is the ribonucleoprotein reverse transcriptase involved in the maintenance of the telomeres, the termini of eukaryotic chromosomes. The RNA component of human telomerase (hTR) consists of 451 nucleotides with the 5' half folding into a highly conserved catalytic core comprising the template region and an adjacent pseudoknot domain (nucleotides 1-208). While the secondary structure of hTR is established, there is little understanding of its three-dimensional (3D) architecture. Here, we have used fluorescence resonance energy transfer (FRET) between fluorescently labelled peptide nucleic acids, hybridized to defined single stranded regions of full length hTR, to evaluate long-range distances. Using molecular modeling, the distance constraints derived by FRET were subsequently used, together with the known secondary structure, to generate a 3D model of the catalytic core of hTR. An overlay of a large set of models generated has provided a low-resolution structure (6.5-8.0 A) that can readily be refined as new structural information becomes available. A notable feature of the modeled structure is the positioning of the template adjacent to the pseudoknot, which brings a number of conserved nucleotides close in space.
Figures



Similar articles
-
Single-molecule FRET-Rosetta reveals RNA structural rearrangements during human telomerase catalysis.RNA. 2017 Feb;23(2):175-188. doi: 10.1261/rna.058743.116. Epub 2016 Nov 15. RNA. 2017. PMID: 28096444 Free PMC article.
-
Analysis of a long-range interaction between conserved domains of human telomerase RNA.RNA. 2004 Jan;10(1):139-47. doi: 10.1261/rna.5118104. RNA. 2004. PMID: 14681592 Free PMC article.
-
Refined secondary-structure models of the core of yeast and human telomerase RNAs directed by SHAPE.RNA. 2015 Feb;21(2):254-61. doi: 10.1261/rna.048959.114. Epub 2014 Dec 15. RNA. 2015. PMID: 25512567 Free PMC article.
-
Regulation of human telomerase RNA biogenesis and localization.RNA Biol. 2021 Mar;18(3):305-315. doi: 10.1080/15476286.2020.1809196. Epub 2020 Sep 2. RNA Biol. 2021. PMID: 32813614 Free PMC article. Review.
-
Structure and function of telomerase RNA.Curr Opin Struct Biol. 2006 Jun;16(3):307-18. doi: 10.1016/j.sbi.2006.05.005. Epub 2006 May 18. Curr Opin Struct Biol. 2006. PMID: 16713250 Review.
Cited by
-
RNA folding pathways from all-atom simulations with a variationally improved history-dependent bias.Biophys J. 2023 Aug 8;122(15):3089-3098. doi: 10.1016/j.bpj.2023.06.012. Epub 2023 Jun 24. Biophys J. 2023. PMID: 37355771 Free PMC article.
-
Single-molecule analysis of telomerase structure and function.Curr Opin Chem Biol. 2011 Dec;15(6):845-52. doi: 10.1016/j.cbpa.2011.10.008. Epub 2011 Nov 5. Curr Opin Chem Biol. 2011. PMID: 22057212 Free PMC article. Review.
-
Oxoisoaporphine as Potent Telomerase Inhibitor.Molecules. 2016 Nov 14;21(11):1534. doi: 10.3390/molecules21111534. Molecules. 2016. PMID: 27854257 Free PMC article.
-
Progress in Human and Tetrahymena Telomerase Structure Determination.Annu Rev Biophys. 2017 May 22;46:199-225. doi: 10.1146/annurev-biophys-062215-011140. Epub 2017 Mar 15. Annu Rev Biophys. 2017. PMID: 28301767 Free PMC article. Review.
-
Three-dimensional RNA structure of the major HIV-1 packaging signal region.Structure. 2013 Jun 4;21(6):951-62. doi: 10.1016/j.str.2013.04.008. Epub 2013 May 16. Structure. 2013. PMID: 23685210 Free PMC article.
References
-
- Weinrich SL, Pruzan R, Ma LB, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, Trager JB, Taylor RD, Carlos R, Andrews WH, Wright WE, Shay JW, Harley CB, Morin GB. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 1997;17:498–502. - PubMed
-
- Beattie TL, Zhou W, Robinson MO, Harrington L. Reconstitution of human telomerase activity in vitro. Curr. Biol. 1998;8:177–180. - PubMed
-
- Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

