Converting a breast cancer microarray signature into a high-throughput diagnostic test
- PMID: 17074082
- PMCID: PMC1636049
- DOI: 10.1186/1471-2164-7-278
Converting a breast cancer microarray signature into a high-throughput diagnostic test
Abstract
Background: A 70-gene tumor expression profile was established as a powerful predictor of disease outcome in young breast cancer patients. This profile, however, was generated on microarrays containing 25,000 60-mer oligonucleotides that are not designed for processing of many samples on a routine basis.
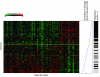
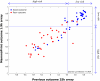
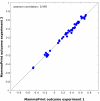
Results: To facilitate its use in a diagnostic setting, the 70-gene prognosis profile was translated into a customized microarray (MammaPrint) containing a reduced set of 1,900 probes suitable for high throughput processing. RNA of 162 patient samples from two previous studies was subjected to hybridization to this custom array to validate the prognostic value. Classification results obtained from the original analysis were then compared to those generated using the algorithms based on the custom microarray and showed an extremely high correlation of prognosis prediction between the original data and those generated using the custom mini-array (p < 0.0001).
Conclusion: In this report we demonstrate for the first time that microarray technology can be used as a reliable diagnostic tool. The data clearly demonstrate the reproducibility and robustness of the small custom-made microarray. The array is therefore an excellent tool to predict outcome of disease in breast cancer patients.
Figures


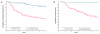
References
-
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. - PubMed
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr., Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling [see comments] Nature. 2000;403:503–511. - PubMed
-
- Perou CM, Sorlie T, Eisen MB, van de RM, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
-
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. - PMC - PubMed
-
- Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, Kim JYH, Goumnerova LC, Black PM, Lau C, Allen JC, Zagzag D, Olson JM, Curran T, Wetmore C, Biegel JA, Poggio T, Mukherjee S, Rifkin R, Califano A, Stolovitzky G, Louis DN, Mesirov JP, Lander ES, Golub TR. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

