Artificially expanded genetic information system: a new base pair with an alternative hydrogen bonding pattern
- PMID: 17074747
- PMCID: PMC1635279
- DOI: 10.1093/nar/gkl633
Artificially expanded genetic information system: a new base pair with an alternative hydrogen bonding pattern
Abstract
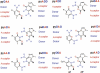
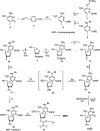
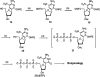
To support efforts to develop a 'synthetic biology' based on an artificially expanded genetic information system (AEGIS), we have developed a route to two components of a non-standard nucleobase pair, the pyrimidine analog 6-amino-5-nitro-3-(1'-beta-D-2'-deoxyribofuranosyl)-2(1H)-pyridone (dZ) and its Watson-Crick complement, the purine analog 2-amino-8-(1'-beta-D-2'-deoxyribofuranosyl)-imidazo[1,2-a]-1,3,5-triazin-4(8H)-one (dP). These implement the pyDDA:puAAD hydrogen bonding pattern (where 'py' indicates a pyrimidine analog and 'pu' indicates a purine analog, while A and D indicate the hydrogen bonding patterns of acceptor and donor groups presented to the complementary nucleobases, from the major to the minor groove). Also described is the synthesis of the triphosphates and protected phosphoramidites of these two nucleosides. We also describe the use of the protected phosphoramidites to synthesize DNA oligonucleotides containing these AEGIS components, verify the absence of epimerization of dZ in those oligonucleotides, and report some hybridization properties of the dZ:dP nucleobase pair, which is rather strong, and the ability of each to effectively discriminate against mismatches in short duplex DNA.
Figures
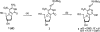

References
-
- Watson J.D., Crick F.H.C. Molecular structure of nucleic acids. Nature. 1953;171:737–738. - PubMed
-
- Watson J.D., Crick F.H.C. General implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. - PubMed
-
- Hentry A.A., Romesberg F.E. Beyond A, C, G, and T: augmenting nature's alphabet. Curr. Opin. Chem. Biol. 2003;7:727–733. - PubMed
-
- Ishikawa M., Hirao I., Yokoyama S. Synthesis of 3-(2-deoxy-β-Dribofuranosyl) pyridin-2-one and 2-amino-6-(N,Ndimethylamino)-9-(2-deoxy-β-D-ribofuranosyl)purine derivatives for an unnatural base pair. Tetrahedron Lett. 2000;41:3931–3934.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

