High-level vancomycin-resistant Staphylococcus aureus isolates associated with a polymicrobial biofilm
- PMID: 17074796
- PMCID: PMC1797660
- DOI: 10.1128/AAC.00576-06
High-level vancomycin-resistant Staphylococcus aureus isolates associated with a polymicrobial biofilm
Abstract
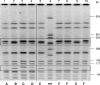
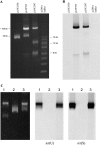
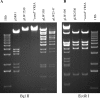
Glycopeptides such as vancomycin are the treatment of choice for infections due to methicillin-resistant Staphylococcus aureus. This study describes the identification of high-level vancomycin-resistant S. aureus (VRSA) isolates in a polymicrobial biofilm within an indwelling nephrostomy tube in a patient in New York. S. aureus, Enterococcus faecalis, Enterococcus faecium, Micrococcus species, Morganella morganii, and Pseudomonas aeruginosa were isolated from the biofilm. For VRSA isolates, vancomycin MICs ranged from 32 to >128 microg/ml. VRSA isolates were also resistant to aminoglycosides, fluoroquinolones, macrolides, penicillin, and tetracycline but remained susceptible to chloramphenicol, linezolid, rifampin, and trimethoprim-sulfamethoxazole. The vanA gene was localized to a plasmid of approximately 100 kb in VRSA and E. faecium isolates from the biofilm. Plasmid analysis revealed that the VRSA isolate acquired the 100-kb E. faecium plasmid, which was then maintained without integration into the MRSA plasmid. The tetracycline resistance genes tet(U) and tet(S), not previously detected in S. aureus isolates, were identified in the VRSA isolates. Additional resistance elements in the VRSA isolate included a multiresistance gene cluster, ermB-aadE-sat4-aphA-3, msrA (macrolide efflux), and the bifunctional aminoglycoside resistance gene aac(6')-aph(2")-Ia. Multiple combinations of resistance genes among the various isolates of staphylococci and enterococci, including vanA, tet(S), and tet(U), illustrate the dynamic nature of gene acquisition and loss within and between bacterial species throughout the course of infection. The potential for interspecies transfer of antimicrobial resistance genes, including resistance to vancomycin, may be enhanced by the microenvironment of a biofilm.
Figures

References
-
- Centers for Disease Control and Prevention. 2002. Staphylococcus aureus resistant to vancomycin—United States, 2002. Morb. Mortal. Wkly. Rep. 51:565-567. - PubMed
-
- Centers for Disease Control and Prevention. 2002. Vancomycin-resistant Staphylococcus aureus—Pennsylvania, 2002. Morb. Mortal. Wkly. Rep. 51:902. - PubMed
-
- Centers for Disease Control and Prevention. 2004. Brief report: vancomycin-resistant Staphylococcus aureus—New York, 2004. Morb. Mortal. Wkly. Rep. 53:322-333. - PubMed
-
- Charpentier, E., G. Gerbaud, and P. Courvalin. 1993. Characterization of a new class of tetracycline-resistance gene tet(S) in Listeria monocytogenes BM4210. Gene 131:27-34. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

