Cardiac myosin binding protein C phosphorylation is cardioprotective
- PMID: 17075052
- PMCID: PMC1636554
- DOI: 10.1073/pnas.0607069103
Cardiac myosin binding protein C phosphorylation is cardioprotective
Abstract
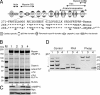
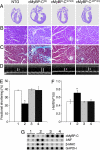
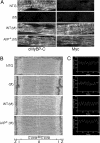
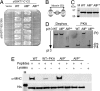
Cardiac myosin binding protein C (cMyBP-C) has three phosphorylatable serines at its N terminus (Ser-273, Ser-282, and Ser-302), and the residues' phosphorylation states may alter thick filament structure and function. To examine the effects of cMyBP-C phosphorylation, we generated transgenic mice with cardiac-specific expression of a cMyBP-C in which the three phosphorylation sites were mutated to aspartic acid, mimicking constitutive phosphorylation (cMyBP-C(AllP+)). The allele was bred into a cMyBP-C null background (cMyBP-C((t/t))) to ensure the absence of endogenous dephosphorylated cMyBP-C. cMyBP-C(AllP+) was incorporated normally into the cardiac sarcomere and restored normal cardiac function in the null background. However, subtle changes in sarcomere ultrastructure, characterized by increased distances between the thick filaments, indicated that phosphomimetic cMyBP-C affects thick-thin filament relationships, and yeast two-hybrid data and pull-down studies both showed that charged residues in these positions effectively prevented interaction with the myosin heavy chain. Confirming the physiological relevance of these data, the cMyBP-C(AllP+:(t/t)) hearts were resistant to ischemia-reperfusion injury. These data demonstrate that cMyBP-C phosphorylation functions in maintaining thick filament spacing and structure and can help protect the myocardium from ischemic injury.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Spirito P, Seidman CE, McKenna WJ, Maron BJ. N Engl J Med. 1997;336:775–785. - PubMed
-
- Bahler M, Moser H, Eppenberger HM, Wallimann T. Dev Biol. 1985;112:345–352. - PubMed
-
- Kawashima M, Kitani S, Tanaka T, Obinata T. J Biochem (Tokyo) 1986;99:1037–1047. - PubMed
-
- Offer G, Moos C, Starr R. J Mol Biol. 1973;74:653–676. - PubMed
-
- Moos C, Mason CM, Besterman JM, Feng IN, Dubin JH. J Mol Biol. 1978;124:571–586. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

