Statistical potential for assessment and prediction of protein structures
- PMID: 17075131
- PMCID: PMC2242414
- DOI: 10.1110/ps.062416606
Statistical potential for assessment and prediction of protein structures
Abstract
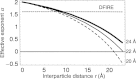
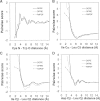
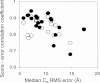
Protein structures in the Protein Data Bank provide a wealth of data about the interactions that determine the native states of proteins. Using the probability theory, we derive an atomic distance-dependent statistical potential from a sample of native structures that does not depend on any adjustable parameters (Discrete Optimized Protein Energy, or DOPE). DOPE is based on an improved reference state that corresponds to noninteracting atoms in a homogeneous sphere with the radius dependent on a sample native structure; it thus accounts for the finite and spherical shape of the native structures. The DOPE potential was extracted from a nonredundant set of 1472 crystallographic structures. We tested DOPE and five other scoring functions by the detection of the native state among six multiple target decoy sets, the correlation between the score and model error, and the identification of the most accurate non-native structure in the decoy set. For all decoy sets, DOPE is the best performing function in terms of all criteria, except for a tie in one criterion for one decoy set. To facilitate its use in various applications, such as model assessment, loop modeling, and fitting into cryo-electron microscopy mass density maps combined with comparative protein structure modeling, DOPE was incorporated into the modeling package MODELLER-8.
Figures



References
-
- Alder, B. 1964. Triplet correlations in hard spheres. Phys. Rev. Lett. 12 317–319.
-
- Anfinsen, C.B. 1973. Principles that govern the folding of protein chains. Science 181 223–230. - PubMed
-
- Bauer, A. and Beyer, A. 1994. An improved pair potential to recognize native protein folds. Proteins 18 254–261. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

