Roles of a Trypanosoma brucei 5'->3' exoribonuclease homolog in mRNA degradation
- PMID: 17077271
- PMCID: PMC1664730
- DOI: 10.1261/rna.291506
Roles of a Trypanosoma brucei 5'->3' exoribonuclease homolog in mRNA degradation
Abstract
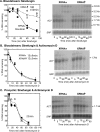
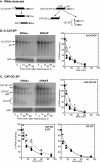
The genome of the kinetoplastid parasite Trypanosoma brucei encodes four homologs of the Saccharomyces cerevisiae 5'-->3' exoribonucleases Xrn1p and Xrn2p/Rat1p, XRNA, XRNB, XRNC, and XRND. In S. cerevisiae, Xrn1p is a cytosolic enzyme involved in degradation of mRNA, whereas Xrn2p is involved in RNA processing in the nucleus. Trypanosome XRND was found in the nucleus, XRNB and XRNC were found in the cytoplasm, and XRNA appeared to be in both compartments. XRND and XRNA were essential for parasite growth. Depletion of XRNA increased the abundances of highly unstable developmentally regulated mRNAs, perhaps by delaying a deadenylation-independent decay pathway. Degradation of more stable or unregulated mRNAs was not affected by XRNA depletion although a slight decrease in average poly(A) tail length was observed. We conclude that in trypanosomes 5'-->3' exonuclease activity is important in degradation of highly unstable, regulated mRNAs, but that for other mRNAs another step is more important in determining the decay rate.
Figures




References
-
- Alibu, P., Storm, L., Haile, S., Clayton, C., Horn, D. A doubly inducible system for RNA interference and rapid RNAi plasmid construction in Trypanosoma brucei . Mol. Biochem. Parasitol. 2004;139:75–82. - PubMed
-
- Amberg, D.C., Goldstein, L.A., Cole, C.N. Isolation and characterization of RAT1: An essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes & Dev. 1992;6:1173–1189. - PubMed
-
- Arraiano, C.M., Maquat, L.E. Post-transcriptional control of gene expression: Effectors of mRNA decay. Mol. Microbiol. 2003;49:267–276. - PubMed
-
- Bashkirov, V.I., Solinger, J.A., Heyer, W.D. Identification of functional domains in the Sep1 protein (= Kem1, Xrn1), which is required for transition through meiotic prophase in Saccharomyces cerevisiae . Chromosoma. 1995;104:215–222. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
