The canonical UPF1-dependent nonsense-mediated mRNA decay is inhibited in transcripts carrying a short open reading frame independent of sequence context
- PMID: 17077274
- PMCID: PMC1664719
- DOI: 10.1261/rna.201406
The canonical UPF1-dependent nonsense-mediated mRNA decay is inhibited in transcripts carrying a short open reading frame independent of sequence context
Abstract
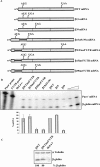
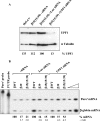
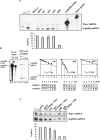
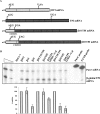
Nonsense-mediated mRNA decay (NMD) is a surveillance mechanism that degrades mRNAs carrying premature translation termination codons. Generally, NMD is elicited if translation terminates >50-54 nucleotides (nt) upstream of an exon-exon junction. We have previously reported that human beta-globin mRNAs carrying 5'-proximal nonsense mutations (e.g., beta15) accumulate to normal levels, suggesting an exception to the "50-54-nt boundary rule." In the present report, we demonstrate that the strength of the UPF1-dependent NMD of mutant beta-globin mRNAs is specifically determined by the proximity of the nonsense codon to the initiation AUG. This conclusion is supported by a parallel effect of the short ORF size on NMD of nonsense-containing alpha-globin mRNAs. To determine whether the short-ORF effect on NMD response is conserved in heterologous transcripts, we assessed its effects on a set of beta-globin/triosephosphate isomerase (TPI) hybrid mRNAs and on the TPI mRNA. Our data support the conclusion that nonsense mutations resulting in a short ORF are able to circumvent the full activity of the canonical UPF1-dependent NMD pathway.
Figures


Similar articles
-
Nonsense mutations in close proximity to the initiation codon fail to trigger full nonsense-mediated mRNA decay.J Biol Chem. 2004 Jul 30;279(31):32170-80. doi: 10.1074/jbc.M405024200. Epub 2004 May 25. J Biol Chem. 2004. PMID: 15161914
-
Resistance of mRNAs with AUG-proximal nonsense mutations to nonsense-mediated decay reflects variables of mRNA structure and translational activity.Nucleic Acids Res. 2015 Jul 27;43(13):6528-44. doi: 10.1093/nar/gkv588. Epub 2015 Jun 11. Nucleic Acids Res. 2015. PMID: 26068473 Free PMC article.
-
Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay.RNA. 2008 Mar;14(3):563-76. doi: 10.1261/rna.815108. Epub 2008 Jan 29. RNA. 2008. PMID: 18230761 Free PMC article.
-
Upf proteins: highly conserved factors involved in nonsense mRNA mediated decay.Mol Biol Rep. 2018 Feb;45(1):39-55. doi: 10.1007/s11033-017-4139-7. Epub 2017 Dec 27. Mol Biol Rep. 2018. PMID: 29282598 Review.
-
Mechanism, factors, and physiological role of nonsense-mediated mRNA decay.Cell Mol Life Sci. 2015 Dec;72(23):4523-44. doi: 10.1007/s00018-015-2017-9. Epub 2015 Aug 18. Cell Mol Life Sci. 2015. PMID: 26283621 Free PMC article. Review.
Cited by
-
Nonsense-mediated RNA decay and its bipolar function in cancer.Mol Cancer. 2021 Apr 29;20(1):72. doi: 10.1186/s12943-021-01364-0. Mol Cancer. 2021. PMID: 33926465 Free PMC article. Review.
-
Ribosomal scanning on the 5'-untranslated region of the human immunodeficiency virus RNA genome.Nucleic Acids Res. 2011 Jul;39(12):5232-44. doi: 10.1093/nar/gkr113. Epub 2011 Mar 9. Nucleic Acids Res. 2011. PMID: 21393254 Free PMC article.
-
Control of human beta-globin mRNA stability and its impact on beta-thalassemia phenotype.Haematologica. 2011 Jun;96(6):905-13. doi: 10.3324/haematol.2010.039206. Epub 2011 Feb 28. Haematologica. 2011. PMID: 21357703 Free PMC article. Review.
-
DMD exon 1 truncating point mutations: amelioration of phenotype by alternative translation initiation in exon 6.Hum Mutat. 2009 Apr;30(4):633-40. doi: 10.1002/humu.20913. Hum Mutat. 2009. PMID: 19206170 Free PMC article.
-
Alternative polyadenylation and nonsense-mediated decay coordinately regulate the human HFE mRNA levels.PLoS One. 2012;7(4):e35461. doi: 10.1371/journal.pone.0035461. Epub 2012 Apr 18. PLoS One. 2012. PMID: 22530027 Free PMC article.
References
-
- Fribourg, S., Gatfield, D., Izaurralde, E., Conti, E. A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat. Struct. Biol. 2003;10:433–439. - PubMed
-
- Gaba, A., Jacobson, A., Sachs, M.S. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol. Cell. 2005;20:449–460. - PubMed
-
- Gehring, N.H., Neu-Yilik, G., Schell, T., Hentze, M.W., Kulozik, A.E. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell. 2003;11:939–949. - PubMed
-
- Ghilardi, N., Skoda, R.C. A single-base deletion in the thrombopoietin (TPO) gene causes familial essential thrombocythemia through a mechanism of more efficient translation of TPO mRNA. Blood. 1999;94:1480–1482. - PubMed
-
- Inácio, A., Silva, A.L., Pinto, J., Ji, X., Morgado, A., Almeida, F., Faustino, P., Lavinha, J., Liebhaber, S.A., Romão, L. Nonsense mutations in close proximity to the initiation codon fail to trigger full nonsense-mediated mRNA decay. J. Biol. Chem. 2004;279:32170–32180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
