Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements
- PMID: 17077319
- PMCID: PMC1665638
- DOI: 10.1101/gr.5565706
Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements
Abstract
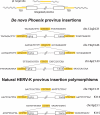
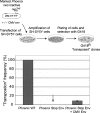
Human Endogenous Retroviruses are expected to be the remnants of ancestral infections of primates by active retroviruses that have thereafter been transmitted in a Mendelian fashion. Here, we derived in silico the sequence of the putative ancestral "progenitor" element of one of the most recently amplified family - the HERV-K family - and constructed it. This element, Phoenix, produces viral particles that disclose all of the structural and functional properties of a bona-fide retrovirus, can infect mammalian, including human, cells, and integrate with the exact signature of the presently found endogenous HERV-K progeny. We also show that this element amplifies via an extracellular pathway involving reinfection, at variance with the non-LTR-retrotransposons (LINEs, SINEs) or LTR-retrotransposons, thus recapitulating ex vivo the molecular events responsible for its dissemination in the host genomes. We also show that in vitro recombinations among present-day human HERV-K (also known as ERVK) loci can similarly generate functional HERV-K elements, indicating that human cells still have the potential to produce infectious retroviruses.
Figures





References
-
- Barbulescu M., Turner G., Seaman M.I., Deinard A.S., Kidd K.K., Lenz J., Turner G., Seaman M.I., Deinard A.S., Kidd K.K., Lenz J., Seaman M.I., Deinard A.S., Kidd K.K., Lenz J., Deinard A.S., Kidd K.K., Lenz J., Kidd K.K., Lenz J., Lenz J. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 1999;9:861–868. - PubMed
-
- Belshaw R., Pereira V., Katzourakis A., Talbot G., Paces J., Burt A., Tristem M., Pereira V., Katzourakis A., Talbot G., Paces J., Burt A., Tristem M., Katzourakis A., Talbot G., Paces J., Burt A., Tristem M., Talbot G., Paces J., Burt A., Tristem M., Paces J., Burt A., Tristem M., Burt A., Tristem M., Tristem M. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. 2004;25:25. - PMC - PubMed
-
- Belshaw R., Dawson A.L., Woolven-Allen J., Redding J., Burt A., Tristem M., Dawson A.L., Woolven-Allen J., Redding J., Burt A., Tristem M., Woolven-Allen J., Redding J., Burt A., Tristem M., Redding J., Burt A., Tristem M., Burt A., Tristem M., Tristem M. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): Implications for present-day activity. J. Virol. 2005;79:12507–12514. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
