Roles of phosphatidate phosphatase enzymes in lipid metabolism
- PMID: 17079146
- PMCID: PMC1769311
- DOI: 10.1016/j.tibs.2006.10.003
Roles of phosphatidate phosphatase enzymes in lipid metabolism
Abstract
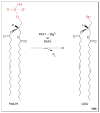
Phosphatidate phosphatase (PAP) enzymes catalyze the dephosphorylation of phosphatidate, yielding diacylglycerol and inorganic phosphate. In eukaryotic cells, PAP activity has a central role in the synthesis of phospholipids and triacylglycerol through its product diacylglycerol, and it also generates and/or degrades lipid-signaling molecules that are related to phosphatidate. There are two types of PAP enzyme, Mg(2+) dependent (PAP1) and Mg(2+) independent (PAP2), but only genes encoding PAP2 enzymes had been identified until recently, when a gene (PAH1) encoding a PAP1 enzyme was found in Saccharomyces cerevisiae. This discovery has revealed a molecular function of the mammalian protein lipin, a deficiency of which causes lipodystrophy in mice. With molecular information now available for both types of PAP, the specific roles of these enzymes in lipid metabolism are being clarified.
Figures

References
-
- Smith SW, et al. The enzymatic dephosphorylation of phosphatidic acids. J Biol Chem. 1957;228:915–922. - PubMed
-
- Jamal Z, et al. Plasma membrane fractions from rat liver contain a phosphatidate phosphohydrolase distinct from that in the endoplasmic reticulum and cytosol. J Biol Chem. 1991;266:2988–2996. - PubMed
-
- Carman GM. Phosphatidate phosphatases and diacylglycerol pyrophosphate phosphatases in Saccharomyces cerevisiae and Escherichia coli. Biochim Biophys Acta. 1997;1348:45–55. - PubMed
-
- Oshiro J, et al. Diacylglycerol pyrophosphate phosphatase in Saccharomyces cerevisiae. Biochim Biophys Acta. 2003;1635:1–9. - PubMed
-
- Sciorra VA, Morris AJ. Roles for lipid phosphate phosphatases in regulation of cellular signaling. Biochim Biophys Acta. 2002;1582:45–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

