Rapid molecular strategy for filovirus detection and characterization
- PMID: 17079496
- PMCID: PMC1828965
- DOI: 10.1128/JCM.01893-06
Rapid molecular strategy for filovirus detection and characterization
Abstract
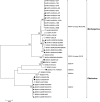
Filoviruses have the capacity to cause lethal outbreaks of hemorrhagic fever in primates. Here we present a simple consensus reverse transcription-PCR method for filovirus recognition and characterization and demonstrate its utility with all known filovirus strains. Phylogenetic assignment is achieved by automated web-based sequence analysis of amplification products.
Figures
References
-
- Domingo, C., G. Palacios, O. Jabado, N. Reyes, M. Niedrig, J. Gascon, M. Cabrerizo, W. I. Lipkin, and A. Tenorio. 2006. Use of a short fragment of the C-terminal E gene for detection and characterization of two new lineages of dengue virus 1 in India. J. Clin. Microbiol. 44:1519-1529. - PMC - PubMed
-
- Hasegawa, M., H. Kishino, and T. Yano. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160-174. - PubMed
-
- Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. - PubMed
-
- Needleman, S. B., and C. D. Wunsch. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48:443-453. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

