The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster
- PMID: 17079687
- PMCID: PMC1620017
- DOI: 10.1101/gad.1482006
The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster
Abstract
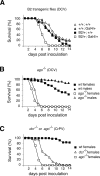
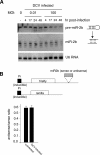
Most organisms have evolved defense mechanisms to protect themselves from viruses and other pathogens. Arthropods lack the protein-based adaptive immune response found in vertebrates. Here we show that the central catalytic component of the RNA-induced silencing complex (RISC), the nuclease Argonaute 2 (Ago-2), is essential for antiviral defense in adult Drosophila melanogaster. Ago-2-defective flies are hypersensitive to infection with a major fruit fly pathogen, Drosophila C virus (DCV), and with Cricket Paralysis virus (CrPV). Increased mortality in ago-2 mutant flies was associated with a dramatic increase in viral RNA accumulation and virus titers. The physiological significance of this antiviral mechanism is underscored by our finding that DCV encodes a potent suppressor of RNA interference (RNAi). This suppressor binds long double-stranded RNA (dsRNA) and inhibits Dicer-2-mediated processing of dsRNA into short interfering RNA (siRNA), but does not bind short siRNAs or disrupt the microRNA (miRNA) pathway. Based on these results we propose that RNAi is a major antiviral immune defense mechanism in Drosophila.
Figures




References
-
- Aravin, A.A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D., Snyder, B., Gaasterland, T., Meyer, J., Tuschl, T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. - PubMed
-
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Baulcombe, D. RNA silencing in plants. Nature. 2004;431:356–363. - PubMed
-
- Bentwich, I., Avniel, A., Karov, Y., Aharonov, R., Gilad, S., Barad, O., Barzilai, A., Einat, P., Einav, U., Meiri, E., et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
