The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages
- PMID: 17079688
- PMCID: PMC1620021
- DOI: 10.1101/gad.1493506
The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages
Abstract
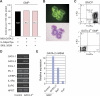
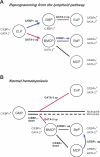
The mechanism of lineage specification in multipotent stem cells has not been fully understood. We recently isolated progenitors with the eosinophil, basophil, or mast cell lineage potential, all of which originate from granulocyte/monocyte progenitors (GMPs). By using these prospectively purified progenitors, we show here that the expression timing of GATA-2 and CCAAT enhancer-binding protein alpha (C/EBPalpha) can differentially control their lineage commitment. The expression of GATA-2 instructed C/EBPalpha-expressing GMPs to commit exclusively into the eosinophil lineage, while it induced basophil and/or mast cell lineage commitment if C/EBPalpha was suppressed at the GMP stage. Furthermore, simply by switching the order of C/EBPalpha and GATA-2 transduction, even lymphoid-committed progenitors recaptured these developmental processes to be reprogrammed into each of these lineages. We propose that the order of expression of key transcription factors is critical for their interplay to selectively drive lineage specification programs, by which stem cells could generate multiple lineage cells in a hierarchical manner.
Figures





References
-
- Akashi, K., Traver, D., Miyamoto, T., Weissman, I.L. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. - PubMed
-
- Brummelkamp, T.R., Bernards, R., Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
