Kidins220/ARMS is transported by a kinesin-1-based mechanism likely to be involved in neuronal differentiation
- PMID: 17079733
- PMCID: PMC1751333
- DOI: 10.1091/mbc.e06-05-0453
Kidins220/ARMS is transported by a kinesin-1-based mechanism likely to be involved in neuronal differentiation
Abstract
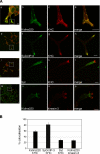
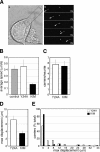
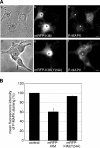
Kinase D-interacting substrate of 220 kDa/ankyrin repeat-rich membrane spanning (Kidins220/ARMS) is a conserved membrane protein mainly expressed in brain and neuroendocrine cells, which is a downstream target of the signaling cascades initiated by neurotrophins and ephrins. We identified kinesin light chain 1 (KLC1) as a binding partner for Kidins220/ARMS by a yeast two-hybrid screen. The interaction between Kidins220/ARMS and the kinesin-1 motor complex was confirmed by glutathione S-transferase-pull-down and coimmunoprecipitation experiments. In addition, Kidins220/ARMS and kinesin-1 were shown to colocalize in nerve growth factor (NGF)-differentiated PC12 cells. Using Kidins220/ARMS and KLC1 mutants, we mapped the regions responsible for the binding to a short sequence of Kidins220/ARMS, termed KLC-interacting motif (KIM), which is sufficient for the interaction with KLC1. Optimal binding of KIM requires a region of KLC1 spanning both the tetratricopeptide repeats and the heptad repeats, previously not involved in cargo recognition. Overexpression of KIM in differentiating PC12 cells impairs the formation and transport of EGFP-Kidins220/ARMS carriers to the tips of growing neurites, leaving other kinesin-1 dependent processes unaffected. Furthermore, KIM overexpression interferes with the activation of the mitogen-activated protein kinase signaling and neurite outgrowth in NGF-treated PC12 cells. Our results suggest that Kidins220/ARMS-positive carriers undergo a kinesin-1-dependent transport linked to neurotrophin action.
Figures





References
-
- Arevalo J. C., Pereira D. B., Yano H., Teng K. K., Chao M. V. Identification of a switch in neurotrophin signaling by selective tyrosine phosphorylation. J. Biol. Chem. 2006;281:1001–1007. - PubMed
-
- Bibel M., Barde Y. A. Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 2000;14:2919–2937. - PubMed
-
- Bohnert S., Schiavo G. Tetanus toxin is transported in a novel neuronal compartment characterized by a specialized pH regulation. J. Biol. Chem. 2005;280:42336–42344. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

