The tree of one percent
- PMID: 17081279
- PMCID: PMC1794558
- DOI: 10.1186/gb-2006-7-10-118
The tree of one percent
Abstract
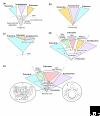
Two significant evolutionary processes are fundamentally not tree-like in nature--lateral gene transfer among prokaryotes and endosymbiotic gene transfer (from organelles) among eukaryotes. To incorporate such processes into the bigger picture of early evolution, biologists need to depart from the preconceived notion that all genomes are related by a single bifurcating tree.
Figures
References
-
- Doolittle WF. If the tree of life fell, would it make a sound? In: Sapp J, editor. Microbial Phylogeny and Evolution: Concepts and Controversies. New York: Oxford University Press; 2004. pp. 119–133.
-
- Mirkin BG, Fenner TI, Galperin MY, Koonin EV. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol Biol. 2003;3:2. doi: 10.1186/1471-2148-3-2. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

