Structural basis for the histone chaperone activity of Asf1
- PMID: 17081973
- PMCID: PMC2981792
- DOI: 10.1016/j.cell.2006.08.047
Structural basis for the histone chaperone activity of Asf1
Abstract
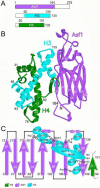
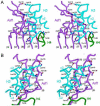
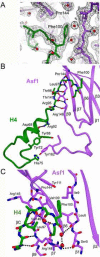
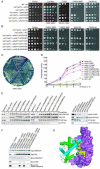
Anti-silencing function 1 (Asf1) is a highly conserved chaperone of histones H3/H4 that assembles or disassembles chromatin during transcription, replication, and repair. The structure of the globular domain of Asf1 bound to H3/H4 determined by X-ray crystallography to a resolution of 1.7 Angstroms shows how Asf1 binds the H3/H4 heterodimer, enveloping the C terminus of histone H3 and physically blocking formation of the H3/H4 heterotetramer. Unexpectedly, the C terminus of histone H4 that forms a mini-beta sheet with histone H2A in the nucleosome undergoes a major conformational change upon binding to Asf1 and adds a beta strand to the Asf1 beta sheet sandwich. Interactions with both H3 and H4 were required for Asf1 histone chaperone function in vivo and in vitro. The Asf1-H3/H4 structure suggests a "strand-capture" mechanism whereby the H4 tail acts as a lever to facilitate chromatin disassembly/assembly that may be used ubiquitously by histone chaperones.
Figures

Comment in
-
Asf1, a loveseat for a histone couple.Cell. 2006 Nov 3;127(3):458-60. doi: 10.1016/j.cell.2006.10.021. Cell. 2006. PMID: 17081967
References
-
- Adkins MW, Howar SR, Tyler JK. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell. 2004;14:657–666. - PubMed
-
- Akey CW, Luger K. Histone chaperones and nucleosome assembly. Curr Opin Struct Biol. 2003;13:6–14. - PubMed
-
- Bailey S. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. - PubMed
-
- Daganzo SM, Erzberger JP, Lam WM, Skordalakes E, Zhang R, Franco AA, Brill SJ, Adams PD, Berger JM, Kaufman PD. Structure and function of the conserved core of histone deposition protein Asf1. Curr Biol. 2003;13:2148–2158. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

