The mechanisms of PML-nuclear body formation
- PMID: 17081985
- PMCID: PMC1978182
- DOI: 10.1016/j.molcel.2006.09.013
The mechanisms of PML-nuclear body formation
Erratum in
-
The Mechanisms of PML-Nuclear Body Formation.Mol Cell. 2006 Dec 8;24(5):805. doi: 10.1016/j.molcel.2006.11.010. Epub 2006 Dec 7. Mol Cell. 2006. PMID: 29654784 No abstract available.
Abstract
PML nuclear bodies (NBs) are nuclear structures that have been implicated in processes such as transcriptional regulation, genome stability, response to viral infection, apoptosis, and tumor suppression. PML has been found to be essential for the formation of the NBs, as these structures do not form in Pml null cells, although PML add back fully rescues their formation. However, the basis for such a structural role of PML is unknown. We demonstrate that PML contains a SUMO binding motif that is independent of its SUMOylation sites and is surprisingly necessary for PML-NB formation. We demonstrate that the PML RING domain is critical for PML SUMOylation and PML-NB formation. We propose a model for PML-NB formation whereby PML SUMOylation and noncovalent binding of PML to SUMOylated PML through the SUMO binding motif constitutes the nucleation event for subsequent recruitment of SUMOylated proteins and/or proteins containing SUMO binding motifs to the PML NBs.
Figures

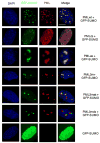
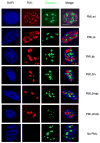
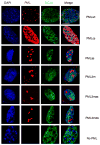


References
-
- Bernardi R, Pandolfi PP. Role of PML and the PML-nuclear body in the control of programmed cell death. Oncogene. 2003;22:9048–9057. - PubMed
-
- Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol 2004 - PubMed
-
- Boddy MN, Howe K, Etkin LD, Solomon E, Freemont PS. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. - PubMed
-
- Dellaire G, Eskiw CH, Dehghani H, Ching RW, Bazett-Jones DP. Mitotic accumulations of PML protein contribute to the re-establishment of PML nuclear bodies in G1. J Cell Sci. 2006;119:1034–1042. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

