U12DB: a database of orthologous U12-type spliceosomal introns
- PMID: 17082203
- PMCID: PMC1635337
- DOI: 10.1093/nar/gkl796
U12DB: a database of orthologous U12-type spliceosomal introns
Abstract
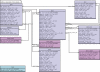
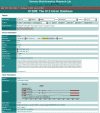
U12-type introns are spliced by the U12-dependent spliceosome and are present in the genomes of many higher eukaryotic lineages including plants, chordates and some invertebrates. However, due to their relatively recent discovery and a systematic bias against recognition of non-canonical splice sites in general, the introns defined by U12-type splice sites are under-represented in genome annotations. Such under-representation compounds the already difficult problem of determining gene structures. It also impedes attempts to study these introns genome-wide or phylum-wide. The resource described here, the U12 Intron Database (U12DB), aims to catalog the U12-type introns of completely sequenced eukaryotic genomes in a framework that groups orthologous introns with each other. This will aid further investigations into the evolution and mechanism of U12-dependent splicing as well as assist ongoing genome annotation efforts. Public access to the U12DB is available at http://genome.imim.es/cgi-bin/u12db/u12db.cgi.
Figures


References
-
- Patel A.A., Steitz J.A. Splicing double: insights from the second spliceosome. Nature Rev. Mol. Cell Biol. 2003;4:960–970. - PubMed
-
- Will C.L., Luhrmann R. Splicing of a rare class of introns by the U12-dependent spliceosome. Biol. Chem. 2005;386:713–724. - PubMed
-
- Hall S.L., Padgett R.A. Conserved sequences in a class of rare eukaryotic nuclear introns with non-consensus splice sites. J. Mol. Biol. 1994;239:357–365. - PubMed

