The evolution of two-component systems in bacteria reveals different strategies for niche adaptation
- PMID: 17083272
- PMCID: PMC1630713
- DOI: 10.1371/journal.pcbi.0020143
The evolution of two-component systems in bacteria reveals different strategies for niche adaptation
Abstract
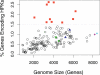
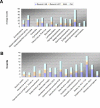
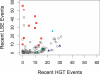
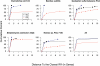
Two-component systems including histidine protein kinases represent the primary signal transduction paradigm in prokaryotic organisms. To understand how these systems adapt to allow organisms to detect niche-specific signals, we analyzed the phylogenetic distribution of nearly 5,000 histidine protein kinases from 207 sequenced prokaryotic genomes. We found that many genomes carry a large repertoire of recently evolved signaling genes, which may reflect selective pressure to adapt to new environmental conditions. Both lineage-specific gene family expansion and horizontal gene transfer play major roles in the introduction of new histidine kinases into genomes; however, there are differences in how these two evolutionary forces act. Genes imported via horizontal transfer are more likely to retain their original functionality as inferred from a similar complement of signaling domains, while gene family expansion accompanied by domain shuffling appears to be a major source of novel genetic diversity. Family expansion is the dominant source of new histidine kinase genes in the genomes most enriched in signaling proteins, and detailed analysis reveals that divergence in domain structure and changes in expression patterns are hallmarks of recent expansions. Finally, while these two modes of gene acquisition are widespread across bacterial taxa, there are clear species-specific preferences for which mode is used.
Conflict of interest statement
Figures

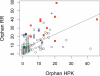
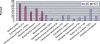



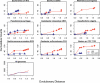
References
-
- Rodrigue A, Quentin Y, Lazdunski A, Mejean V, Foglino M. Two-component systems in Pseudomonas aeruginosa: Why so many? Trends Microbiol. 2000;8:498–504. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

