Transcriptome changes in the phenylpropanoid pathway of Glycine max in response to Pseudomonas syringae infection
- PMID: 17083738
- PMCID: PMC1636052
- DOI: 10.1186/1471-2229-6-26
Transcriptome changes in the phenylpropanoid pathway of Glycine max in response to Pseudomonas syringae infection
Abstract
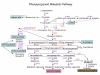
Background: Reports of plant molecular responses to pathogenic infections have pinpointed increases in activity of several genes of the phenylpropanoid pathway leading to the synthesis of lignin and flavonoids. The majority of those findings were derived from single gene studies and more recently from several global gene expression analyses. We undertook a global transcriptional analysis focused on the response of genes of the multiple branches of the phenylpropanoid pathway to infection by the Pseudomonas syringae pv. glycinea with or without the avirulence gene avrB to characterize more broadly the contribution of the multiple branches of the pathway to the resistance response in soybean. Transcript abundance in leaves was determined from analysis of soybean cDNA microarray data and hybridizations to RNA blots with specific gene probes.
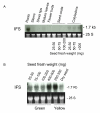
Results: The majority of the genes surveyed presented patterns of increased transcript accumulation. Some increased rapidly, 2 and 4 hours after inoculation, while others started to accumulate slowly by 8-12 hours. In contrast, transcripts of a few genes decreased in abundance 2 hours post inoculation. Most interestingly was the opposite temporal fluctuation in transcript abundance between early responsive genes in defense (CHS and IFS1) and F3H, the gene encoding a pivotal enzyme in the synthesis of anthocyanins, proanthocyanidins and flavonols. F3H transcripts decreased rapidly 2 hours post inoculation and increased during periods when CHS and IFS transcripts decreased. It was also determined that all but one (CHS4) family member genes (CHS1, CHS2, CHS3, CHS5, CHS6 and CHS7/8) accumulated higher transcript levels during the defense response provoked by the avirulent pathogen challenge.
Conclusion: Based on the mRNA profiles, these results show the strong bias that soybean has towards increasing the synthesis of isoflavonoid phytoalexins concomitant with the down regulation of genes required for the synthesis of anthocyanins and proanthocyanins. Although proanthocyanins are known to be toxic compounds, the cells in the soybean leaves seem to be programmed to prioritize the synthesis and accumulation of isoflavonoid and pterocarpan phytoalexins during the resistance response. It was known that CHS transcripts accumulate in great abundance rapidly after inoculation of the soybean plants but our results have demonstrated that all but one (CHS4) member of the gene family member genes accumulated higher transcript levels during the defense response.
Figures






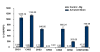
References
-
- Cross JE, Kennedy BW, Lambert JW, Cooper RL. Pathogenic races of the bacterial pathogen of soybeans, Pseudomonas glycinea. Plant Disease Report. 1966;50:557–560.
-
- Klement Z. From hypersensitivity defense reaction of plants to pathogens. In: Mount MS, Lacy GH, editor. Phytopathogenic Prokariotes. New York: Academic Press; 1982. pp. 149–177.
-
- Jones AM, Dangl JL. Logjam at the Styx: Programed Cell Death in Plants. Trends in Plant Sciences. 1996;1:114–119. doi: 10.1016/S1360-1385(96)90005-9. - DOI
-
- de Wit PJGM. Pathogen avirulence and plant resistance: A key role for recognition. Trends Plant Sci. 1997;2:452–458. doi: 10.1016/S1360-1385(97)01139-4. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

