Transcriptional regulation of the FSH receptor: new perspectives
- PMID: 17084019
- PMCID: PMC3682414
- DOI: 10.1016/j.mce.2006.09.005
Transcriptional regulation of the FSH receptor: new perspectives
Abstract
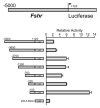
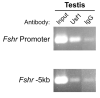
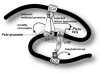
The cell-surface receptor for the gonadotropin follicle-stimulating hormone (FSH) is expressed exclusively on Sertoli cells of the testis and granulosa cells of the ovary. FSH signal transduction through its receptor (Fshr) is critical for the timing and maintenance of normal gametogenesis in the mammalian gonad. In the 13 years since the gene encoding Fshr was first cloned, the mechanisms controlling its transcription have been extensively examined, but a clear understanding of what drives its unique cell-specificity remains elusive. Current knowledge of basal Fshr transcription highlights the role of an E-box in the proximal promoter which is bound by the basic helix-loop-helix transcription factors upstream stimulatory factor 1 (Usf1) and Usf2. Recent studies utilizing knockout mice and chromatin immunoprecipitation validated the importance of Usf to Fshr transcription and demonstrated a sexually dimorphic requirement for the Usf proteins to maintain normal Fshr expression. Studies have also shown that the promoter region itself is insufficient for appropriate Fshr expression in transgenic mice, indicating Fshr transcription depends on regulatory elements that lie outside of the promoter. Identification of such elements has been propelled by recent availability of genome sequence data, which facilitated studies using comparative genomics, DNase I hypersensitivity mapping, and transgenic analysis with large fragments of DNA. This review will focus on the current understanding of transcriptional regulatory processes that control expression of rat Fshr, including recent advances from our laboratory.
Figures



Similar articles
-
Follicle-stimulating hormone (FSH) transiently blocks FSH receptor transcription by increasing inhibitor of deoxyribonucleic acid binding/differentiation-2 and decreasing upstream stimulatory factor expression in rat Sertoli cells.Endocrinology. 2009 Aug;150(8):3783-91. doi: 10.1210/en.2008-1261. Epub 2009 May 7. Endocrinology. 2009. PMID: 19423764 Free PMC article.
-
In vivo regulation of follicle-stimulating hormone receptor by the transcription factors upstream stimulatory factor 1 and upstream stimulatory factor 2 is cell specific.Endocrinology. 2008 Oct;149(10):5297-306. doi: 10.1210/en.2007-1199. Epub 2008 Jun 19. Endocrinology. 2008. PMID: 18566134 Free PMC article.
-
The USF proteins regulate transcription of the follicle-stimulating hormone receptor but are insufficient for cell-specific expression.Mol Endocrinol. 2000 Nov;14(11):1836-48. doi: 10.1210/mend.14.11.0557. Mol Endocrinol. 2000. PMID: 11075816 Free PMC article.
-
Current concepts of follicle-stimulating hormone receptor gene regulation.Biol Reprod. 2011 Jan;84(1):7-17. doi: 10.1095/biolreprod.110.085043. Epub 2010 Aug 25. Biol Reprod. 2011. PMID: 20739665 Free PMC article. Review.
-
The expression of the follicle-stimulating hormone receptor in spermatogenesis.Recent Prog Horm Res. 2002;57:129-48. doi: 10.1210/rp.57.1.129. Recent Prog Horm Res. 2002. PMID: 12017540 Free PMC article. Review.
Cited by
-
De novo transcriptome sequencing and analysis of male and female swimming crab (Portunus trituberculatus) reproductive systems during mating embrace (stage II).BMC Genet. 2018 Jan 3;19(1):3. doi: 10.1186/s12863-017-0592-5. BMC Genet. 2018. PMID: 29298661 Free PMC article.
-
Upstream stimulatory factor induces Nr5a1 and Shbg gene expression during the onset of rat Sertoli cell differentiation.Biol Reprod. 2011 Nov;85(5):965-76. doi: 10.1095/biolreprod.111.093013. Epub 2011 Jul 6. Biol Reprod. 2011. PMID: 21734262 Free PMC article.
-
Follicle-stimulating hormone (FSH) transiently blocks FSH receptor transcription by increasing inhibitor of deoxyribonucleic acid binding/differentiation-2 and decreasing upstream stimulatory factor expression in rat Sertoli cells.Endocrinology. 2009 Aug;150(8):3783-91. doi: 10.1210/en.2008-1261. Epub 2009 May 7. Endocrinology. 2009. PMID: 19423764 Free PMC article.
-
Dual effects of gonadotropin-inhibitory hormone on testicular development in prepubertal Minxinan Black rabbits (Oryctolagus cuniculus).Front Vet Sci. 2024 Jan 24;11:1320452. doi: 10.3389/fvets.2024.1320452. eCollection 2024. Front Vet Sci. 2024. PMID: 38328257 Free PMC article.
-
Association of single-nucleotide polymorphisms in the ESR2 and FSHR genes with poor ovarian response in infertile Jordanian women.Clin Exp Reprod Med. 2021 Mar;48(1):69-79. doi: 10.5653/cerm.2020.03706. Epub 2021 Jan 28. Clin Exp Reprod Med. 2021. PMID: 33503363 Free PMC article.
References
-
- Bishop CE, Whitworth DJ, Qin Y, Agoulnik AI, Agoulnik IU, Harrison WR, Behringer RR, Overbeek PA. A transgenic insertion upstream of Sox9 is associated with dominant XX sex reversal in the mouse. Nat.Genet. 2000;26:490–494. - PubMed
-
- Bluhm AP, Toledo RA, Mesquita FM, Pimenta MT, Fernandes FM, Ribela MT, Lazari MF. Molecular cloning, sequence analysis and expression of the snake Follicle-stimulating hormone receptor. Gen.Comp Endocrinol. 2004;137:300–311. - PubMed
-
- Bockers TM, Nieschlag E, Kreutz MR, Bergmann M. Localization of Follicle-stimulating hormone (FSH) immunoreactivity and hormone receptor mRNA in testicular tissue of infertile men. Cell Tissue Res. 1994;278:595–600. - PubMed
-
- Camp TA, Rahal JO, Mayo KE. Cellular localization and hormonal regulation of Follicle-stimulating hormone and Luteinizaing hormone receptor messenger RNAs in the rat ovary. Mol.Endocrinol. 1991;5:1405–1417. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

