Variation of enzyme activities and metabolite levels in 24 Arabidopsis accessions growing in carbon-limited conditions
- PMID: 17085515
- PMCID: PMC1676042
- DOI: 10.1104/pp.106.086629
Variation of enzyme activities and metabolite levels in 24 Arabidopsis accessions growing in carbon-limited conditions
Abstract
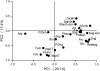
Our understanding of the interaction of carbon (C) metabolism with nitrogen (N) metabolism and growth is based mainly on studies of responses to environmental treatments, and studies of mutants and transformants. Here, we investigate which metabolic parameters vary and which parameters change in a coordinated manner in 24 genetically diverse Arabidopsis (Arabidopsis thaliana) accessions, grown in C-limited conditions. The accessions were grown in short days, moderate light, and high nitrate, and analyzed for rosette biomass, levels of structural components (protein, chlorophyll), total phenols and major metabolic intermediates (sugars, starch, nitrate, amino acids), and the activities of seven representative enzymes from central C and N metabolism. The largest variation was found for plant weight, reducing sugars, starch at the end of the night, and several enzyme activities. High levels of one sugar correlated with high levels of other sugars and starch, and a trend to increased amino acids, slightly lower nitrate, and higher protein. The activities of enzymes at the interface of C and N metabolism correlated with each other, but were unrelated to carbohydrates, amino acid levels, and total protein. Rosette weight was unrelated or showed a weak negative trend to sugar and amino acid contents at the end of the day in most of the accessions, and was negatively correlated with starch at the end of the night. Rosette weight was positively correlated with several enzyme activities. We propose that growth is not related to the absolute levels of starch, sugars, and amino acids; instead, it is related to flux, which is indicated by the enzymatic capacity to use these central resources.
Figures




References
-
- Bantan-Polak T, Kassai M, Grant KB (2001) A comparison of fluorescamine and naphthalene-2,3-dicarboxaldehyde fluorogenic reagents for microplate-based detection of amino acids. Anal Biochem 297: 128–136 - PubMed
-
- Causse M, Rocher JP, Henry AM, Charcosset A, Prioul JL, de Vienne D (1995) Genetic dissection of the relationship between carbon metabolism and early growth in maize, with emphasis on key enzyme loci. Mol Breed 1: 259–272
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

