A new look at bacteriophage lambda genetic networks
- PMID: 17085553
- PMCID: PMC1797383
- DOI: 10.1128/JB.01215-06
A new look at bacteriophage lambda genetic networks
Figures

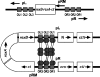
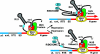
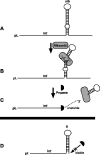
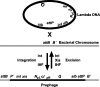
References
-
- Aurell, E., S. Brown, J. Johanson, and K. Sneppen. 2002. Stability puzzles in phage lambda. Phys. Rev. E 65:051914-051919. - PubMed
-
- Banuett, F., M. A. Hoyt, L. McFarlane, H. Echols, and I. Herskowitz. 1986. hflB, a new Escherichia coli locus regulating lysogeny and the level of bacteriophage lambda cII protein. J. Mol. Biol. 187:213-224. - PubMed
-
- Britton, R. A., B. S. Powell, S. Dasgupta, Q. Sun, W. Margolin, J. R. Lupski, and D. L. Court. 1998. Cell cycle arrest in Era GTPase mutants: a potential growth rate-regulated checkpoint in Escherichia coli. Mol. Microbiol. 27:739-750. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

