Characterization of a higBA toxin-antitoxin locus in Vibrio cholerae
- PMID: 17085558
- PMCID: PMC1797405
- DOI: 10.1128/JB.00909-06
Characterization of a higBA toxin-antitoxin locus in Vibrio cholerae
Abstract
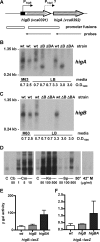
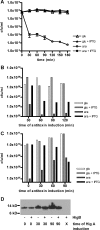
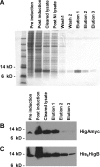
Toxin-antitoxin (TA) loci, which were initially characterized as plasmid stabilization agents, have in recent years been detected on the chromosomes of numerous free-living bacteria. Vibrio cholerae, the causative agent of cholera, contains 13 putative TA loci, all of which are clustered within the superintegron on chromosome II. Here we report the characterization of the V. cholerae higBA locus, also known as VCA0391/2. Deletion of higA alone was not possible, consistent with predictions that it encodes an antitoxin, and biochemical analyses confirmed that HigA interacts with HigB. Transient exogenous expression of the toxin HigB dramatically slowed growth of V. cholerae and Escherichia coli and reduced the numbers of CFU by several orders of magnitude. HigB toxicity could be counteracted by simultaneous or delayed production of HigA, although HigA's effect diminished as the delay lengthened. Transcripts from endogenous higBA increased following treatment of V. cholerae with translational inhibitors, presumably due to reduced levels of HigA, which represses the higBA locus. However, no higBA-dependent cell death was observed in response to such stimuli. Thus, at least under the conditions tested, activation of endogenous HigB does not appear to be bactericidal.
Figures

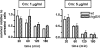
References
-
- Buts, L., J. Lah, M. H. Dao-Thi, L. Wyns, and R. Loris. 2005. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem. Sci. 30:672-679. - PubMed
-
- Christensen, S. K., and K. Gerdes. 2003. RelE toxins from bacteria and archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol. Microbiol. 48:1389-1400. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

