A simple spreadsheet-based, MIAME-supportive format for microarray data: MAGE-TAB
- PMID: 17087822
- PMCID: PMC1687205
- DOI: 10.1186/1471-2105-7-489
A simple spreadsheet-based, MIAME-supportive format for microarray data: MAGE-TAB
Abstract
Background: Sharing of microarray data within the research community has been greatly facilitated by the development of the disclosure and communication standards MIAME and MAGE-ML by the MGED Society. However, the complexity of the MAGE-ML format has made its use impractical for laboratories lacking dedicated bioinformatics support.
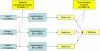
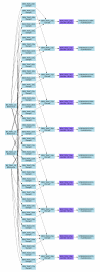
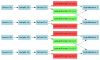
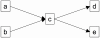
Results: We propose a simple tab-delimited, spreadsheet-based format, MAGE-TAB, which will become a part of the MAGE microarray data standard and can be used for annotating and communicating microarray data in a MIAME compliant fashion.
Conclusion: MAGE-TAB will enable laboratories without bioinformatics experience or support to manage, exchange and submit well-annotated microarray data in a standard format using a spreadsheet. The MAGE-TAB format is self-contained, and does not require an understanding of MAGE-ML or XML.
Figures


References
-
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. - DOI - PubMed
-
- Spellman PT, Miller M, Stewart J, Troup C, Sarkans U, Chervitz S, Bernhart D, Sherlock G, Ball C, Lepage M, Swiatek M, Marks WL, Goncalves J, Markel S, Iordan D, Shojatalab M, Pizarro A, White J, Hubley R, Deutsch E, Senger M, Aronow BJ, Robinson A, Bassett D, Stoeckert CJ, Brazma A. Design and implementation of microarray gene expression markup language (MAGE-ML) Genome Biol. 2002;3:RESEARCH0046. doi: 10.1186/gb-2002-3-9-research0046. - DOI - PMC - PubMed
-
- Parkinson H, Sarkans U, Shojatalab M, Abeygunawardena N, Contrino S, Coulson R, Farne A, Lara GG, Holloway E, Kapushesky M, Lilja P, Mukherjee G, Oezcimen A, Rayner T, Rocca-Serra P, Sharma A, Sansone S, Brazma A. ArrayExpress – a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2005:D553–555. - PMC - PubMed
-
- Ball CA, Brazma A, Causton H, Chervitz S, Edgar R, Hingamp P, Matese JC, Parkinson H, Quackenbush J, Ringwald M, Sansone SA, Sherlock G, Spellman P, Stoeckert C, Tateno Y, Taylor R, White J, Winegarden N. Submission of microarray data to public repositories. PLoS Biol. 2004;2:E317. doi: 10.1371/journal.pbio.0020317. - DOI - PMC - PubMed
-
- Microarray standards at last. Nature. 2002;419:323. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

