Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression
- PMID: 17088209
- PMCID: PMC1713190
- DOI: 10.1016/j.neuron.2006.08.028
Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression
Abstract
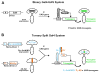
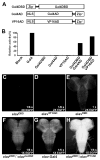
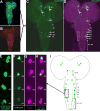
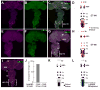
Selective genetic manipulation of neuronal function in vivo requires techniques for targeting gene expression to specific cells. Existing systems accomplish this using the promoters of endogenous genes to drive expression of transgenes directly in cells of interest or, in "binary" systems, to drive expression of a transcription factor or recombinase that subsequently activates the expression of other transgenes. All such techniques are constrained by the limited specificity of the available promoters. We introduce here a combinatorial system in which the DNA-binding (DBD) and transcription-activation (AD) domains of a transcription factor are independently targeted using two different promoters. The domains heterodimerize to become transcriptionally competent and thus drive transgene expression only at the intersection of the expression patterns of the two promoters. We use this system to dissect a neuronal network in Drosophila by selectively targeting expression of the cell death gene reaper to subsets of neurons within the network.
Figures




Similar articles
-
GAL4-NF-kappaB fusion protein augments transgene expression from neuronal promoters in the rat brain.Mol Ther. 2006 Dec;14(6):872-82. doi: 10.1016/j.ymthe.2006.05.020. Epub 2006 Aug 10. Mol Ther. 2006. PMID: 16904943
-
Combinatorial methods for refined neuronal gene targeting.Curr Opin Neurobiol. 2007 Oct;17(5):572-80. doi: 10.1016/j.conb.2007.10.001. Epub 2007 Nov 19. Curr Opin Neurobiol. 2007. PMID: 18024005 Review.
-
Characterization of Drosophila fruitless-gal4 transgenes reveals expression in male-specific fruitless neurons and innervation of male reproductive structures.J Comp Neurol. 2004 Jul 19;475(2):270-87. doi: 10.1002/cne.20177. J Comp Neurol. 2004. PMID: 15211467
-
An efficient system for tissue-specific overexpression of transgenes in podocytes in vivo.Am J Physiol Renal Physiol. 2005 Aug;289(2):F481-8. doi: 10.1152/ajprenal.00332.2004. Epub 2005 Mar 22. Am J Physiol Renal Physiol. 2005. PMID: 15784842
-
The GAL4 System: A Versatile System for the Manipulation and Analysis of Gene Expression.Methods Mol Biol. 2016;1478:33-52. doi: 10.1007/978-1-4939-6371-3_2. Methods Mol Biol. 2016. PMID: 27730574 Review.
Cited by
-
Light, heat, action: neural control of fruit fly behaviour.Philos Trans R Soc Lond B Biol Sci. 2015 Sep 19;370(1677):20140211. doi: 10.1098/rstb.2014.0211. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26240426 Free PMC article. Review.
-
Imaging neural activity in the ventral nerve cord of behaving adult Drosophila.Nat Commun. 2018 Oct 22;9(1):4390. doi: 10.1038/s41467-018-06857-z. Nat Commun. 2018. PMID: 30348941 Free PMC article.
-
A dedicate sensorimotor circuit enables fine texture discrimination by active touch.PLoS Genet. 2023 Jan 17;19(1):e1010562. doi: 10.1371/journal.pgen.1010562. eCollection 2023 Jan. PLoS Genet. 2023. PMID: 36649336 Free PMC article.
-
The utility and caveat of split-GAL4s in the study of neurodegeneration.Fly (Austin). 2023 Dec;17(1):2192847. doi: 10.1080/19336934.2023.2192847. Fly (Austin). 2023. PMID: 36959085 Free PMC article.
-
A Genetic Toolkit for Dissecting Dopamine Circuit Function in Drosophila.Cell Rep. 2018 Apr 10;23(2):652-665. doi: 10.1016/j.celrep.2018.03.068. Cell Rep. 2018. PMID: 29642019 Free PMC article.
References
-
- Awatramani R, Soriano P, Rodriguez C, Mai JJ, Dymecki SM. Cryptic boundaries in roof plate and choroid plexus identified by intersectional gene activation. Nat Genet. 2003;35:70–75. - PubMed
-
- Bellen HJ, O'Kane CJ, Wilson C, Grossniklaus U, Pearson RK, Gehring WJ. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 1989;3:1288–1300. - PubMed
-
- Brand AH, Perrimon N. Targeted Gene-Expression As A Means Of Altering Cell Fates And Generating Dominant Phenotypes. Development. 1993;118:401–415. - PubMed
-
- Brent R, Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985;43:729–736. - PubMed
-
- Cahill MA, Ernst WH, Janknecht R, Nordheim A. Regulatory Squelching. Febs Letters. 1994;344:105–108. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

