The Escherichia coli argW-dsdCXA genetic island is highly variable, and E. coli K1 strains commonly possess two copies of dsdCXA
- PMID: 17088369
- PMCID: PMC1698345
- DOI: 10.1128/JCM.01172-06
The Escherichia coli argW-dsdCXA genetic island is highly variable, and E. coli K1 strains commonly possess two copies of dsdCXA
Abstract
The genome sequences of Escherichia coli pathotypes reveal extensive genetic variability in the argW-dsdCXA island. Interestingly, the archetype E. coli K1 neonatal meningitis strain, strain RS218, has two copies of the dsdCXA genes for d-serine utilization at the argW and leuX islands. Because the human brain contains d-serine, an epidemiological study emphasizing K1 isolates surveyed the dsdCXA copy number and function. Forty of 41 (97.5%) independent E. coli K1 isolates could utilize d-serine. Southern blot hybridization revealed physical variability within the argW-dsdC region, even among 22 E. coli O18:K1:H7 isolates. In addition, 30 of 41 K1 strains, including 21 of 22 O18:K1:H7 isolates, had two dsdCXA loci. Mutational analysis indicated that each of the dsdA genes is functional in a rifampin-resistant mutant of RS218, mutant E44. The high percentage of K1 strains that can use d-serine is in striking contrast to our previous observation that only 4 of 74 (5%) isolates in the diarrheagenic E. coli (DEC) collection have this activity. The genome sequence of diarrheagenic E. coli isolates indicates that the csrRAKB genes for sucrose utilization are often substituted for dsdC and a portion of dsdX present at the argW-dsdCXA island of extraintestinal isolates. Among DEC isolates there is a reciprocal pattern of sucrose fermentation versus d-serine utilization. The ability to use d-serine is a trait strongly selected for among E. coli K1 strains, which have the ability to infect a wide range of extraintestinal sites. Conversely, diarrheagenic E. coli pathotypes appear to have substituted sucrose for d-serine as a potential nutrient.
Figures

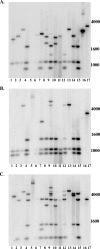

Similar articles
-
DsdX is the second D-serine transporter in uropathogenic Escherichia coli clinical isolate CFT073.J Bacteriol. 2006 Sep;188(18):6622-8. doi: 10.1128/JB.00634-06. J Bacteriol. 2006. PMID: 16952954 Free PMC article.
-
Roles of serine accumulation and catabolism in the colonization of the murine urinary tract by Escherichia coli CFT073.Infect Immun. 2007 Nov;75(11):5298-304. doi: 10.1128/IAI.00652-07. Epub 2007 Sep 4. Infect Immun. 2007. PMID: 17785472 Free PMC article.
-
d-Serine induces distinct transcriptomes in diverse Escherichia coli pathotypes.Microbiology (Reading). 2021 Oct;167(10). doi: 10.1099/mic.0.001093. Microbiology (Reading). 2021. PMID: 34623236
-
Mobile contingency locus controlling Escherichia coli K1 polysialic acid capsule acetylation.Mol Microbiol. 2006 May;60(4):828-37. doi: 10.1111/j.1365-2958.2006.05158.x. Mol Microbiol. 2006. PMID: 16677296 Review.
-
Molecular epidemiology of Escherichia coli causing neonatal meningitis.Int J Med Microbiol. 2005 Oct;295(6-7):373-81. doi: 10.1016/j.ijmm.2005.07.011. Int J Med Microbiol. 2005. PMID: 16238014 Review.
Cited by
-
Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli.J Bacteriol. 2007 May;189(9):3532-46. doi: 10.1128/JB.01744-06. Epub 2007 Mar 9. J Bacteriol. 2007. PMID: 17351047 Free PMC article.
-
The Scr and Csc pathways for sucrose utilization co-exist in E. coli, but only the Scr pathway is widespread in other Enterobacteriaceae.Front Microbiol. 2024 Jul 3;15:1409295. doi: 10.3389/fmicb.2024.1409295. eCollection 2024. Front Microbiol. 2024. PMID: 39021635 Free PMC article.
-
The genome sequence of E. coli W (ATCC 9637): comparative genome analysis and an improved genome-scale reconstruction of E. coli.BMC Genomics. 2011 Jan 6;12:9. doi: 10.1186/1471-2164-12-9. BMC Genomics. 2011. PMID: 21208457 Free PMC article.
-
Genomic and Phenomic Study of Mammary Pathogenic Escherichia coli.PLoS One. 2015 Sep 1;10(9):e0136387. doi: 10.1371/journal.pone.0136387. eCollection 2015. PLoS One. 2015. PMID: 26327312 Free PMC article.
-
Revisiting the host as a growth medium.Nat Rev Microbiol. 2008 Sep;6(9):657-66. doi: 10.1038/nrmicro1955. Nat Rev Microbiol. 2008. PMID: 18679171 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

