The RNA polymerase II kinase Ctk1 regulates positioning of a 5' histone methylation boundary along genes
- PMID: 17088384
- PMCID: PMC1800795
- DOI: 10.1128/MCB.01628-06
The RNA polymerase II kinase Ctk1 regulates positioning of a 5' histone methylation boundary along genes
Abstract
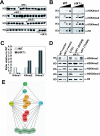
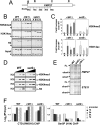
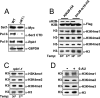
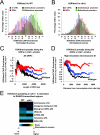
In yeast and other eukaryotes, the histone methyltransferase Set1 mediates methylation of lysine 4 on histone H3 (H3K4me). This modification marks the 5' end of transcribed genes in a 5'-to-3' tri- to di- to monomethyl gradient and promotes association of chromatin-remodeling and histone-modifying enzymes. Here we show that Ctk1, the serine 2 C-terminal domain (CTD) kinase for RNA polymerase II (RNAP II), regulates H3K4 methylation. We found that CTK1 deletion nearly abolished H3K4 monomethylation yet caused a significant increase in H3K4 di- and trimethylation. Both in individual genes and genome-wide, loss of CTK1 disrupted the H3K4 methylation patterns normally observed. H3K4me2 and H3K4me3 spread 3' into the bodies of genes, while H3K4 monomethylation was diminished. These effects were dependent on the catalytic activity of Ctk1 but are independent of Set2-mediated H3K36 methylation. Furthermore, these effects are not due to spurious transcription initiation in the bodies of genes, to changes in RNAP II occupancy, to changes in serine 5 CTD phosphorylation patterns, or to "transcriptional stress." These data show that Ctk1 acts to restrict the spread of H3K4 methylation through a mechanism that is independent of a general transcription defect. The evidence presented suggests that Ctk1 controls the maintenance of suppressive chromatin in the coding regions of genes by both promoting H3K36 methylation, which leads to histone deacetylation, and preventing the 3' spread of H3K4 trimethylation, a mark associated with transcriptional initiation.
Figures

Similar articles
-
Ctk complex-mediated regulation of histone methylation by COMPASS.Mol Cell Biol. 2007 Jan;27(2):709-20. doi: 10.1128/MCB.01627-06. Epub 2006 Nov 6. Mol Cell Biol. 2007. PMID: 17088385 Free PMC article.
-
Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36.Mol Cell Biol. 2008 Aug;28(16):4915-26. doi: 10.1128/MCB.00001-08. Epub 2008 Jun 9. Mol Cell Biol. 2008. PMID: 18541663 Free PMC article.
-
DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes.PLoS Genet. 2010 Oct 28;6(10):e1001173. doi: 10.1371/journal.pgen.1001173. PLoS Genet. 2010. PMID: 21060864 Free PMC article.
-
Diverse and dynamic forms of gene regulation by the S. cerevisiae histone methyltransferase Set1.Curr Genet. 2023 Jun;69(2-3):91-114. doi: 10.1007/s00294-023-01265-3. Epub 2023 Mar 31. Curr Genet. 2023. PMID: 37000206 Review.
-
Context-Dependent and Locus-Specific Role of H3K36 Methylation in Transcriptional Regulation.J Mol Biol. 2025 Jan 1;437(1):168796. doi: 10.1016/j.jmb.2024.168796. Epub 2024 Sep 19. J Mol Biol. 2025. PMID: 39299382 Review.
Cited by
-
Sus1p facilitates pre-initiation complex formation at the SAGA-regulated genes independently of histone H2B de-ubiquitylation.J Mol Biol. 2014 Aug 12;426(16):2928-2941. doi: 10.1016/j.jmb.2014.05.028. Epub 2014 Jun 6. J Mol Biol. 2014. PMID: 24911582 Free PMC article.
-
Human TFIIH Kinase CDK7 Regulates Transcription-Associated Chromatin Modifications.Cell Rep. 2017 Aug 1;20(5):1173-1186. doi: 10.1016/j.celrep.2017.07.021. Cell Rep. 2017. PMID: 28768201 Free PMC article.
-
In vivo effects of histone H3 depletion on nucleosome occupancy and position in Saccharomyces cerevisiae.PLoS Genet. 2012;8(6):e1002771. doi: 10.1371/journal.pgen.1002771. Epub 2012 Jun 21. PLoS Genet. 2012. PMID: 22737086 Free PMC article.
-
Knockdown of SLBP results in nuclear retention of histone mRNA.RNA. 2009 Mar;15(3):459-72. doi: 10.1261/rna.1205409. Epub 2009 Jan 20. RNA. 2009. PMID: 19155325 Free PMC article.
-
The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis.Annu Rev Biochem. 2012;81:65-95. doi: 10.1146/annurev-biochem-051710-134100. Annu Rev Biochem. 2012. PMID: 22663077 Free PMC article. Review.
References
-
- Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13:67-76. - PubMed
-
- Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. - PubMed
-
- Bohlander, S. K., R. Espinosa III, M. M. Le Beau, J. D. Rowley, and M. O. Diaz. 1992. A method for the rapid sequence-independent amplification of microdissected chromosomal material. Genomics 13:1322-1324. - PubMed
-
- Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson, K. K. Lee, W. J. Shia, S. Anderson, J. Yates, M. P. Washburn, and J. L. Workman. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581-592. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
