How many metals does it take to fix N2? A mechanistic overview of biological nitrogen fixation
- PMID: 17088547
- PMCID: PMC1859894
- DOI: 10.1073/pnas.0603978103
How many metals does it take to fix N2? A mechanistic overview of biological nitrogen fixation
Abstract
During the process of biological nitrogen fixation, the enzyme nitrogenase catalyzes the ATP-dependent reduction of dinitrogen to ammonia. Nitrogenase consists of two component metalloproteins, the iron (Fe) protein and the molybdenum-iron (MoFe) protein; the Fe protein mediates the coupling of ATP hydrolysis to interprotein electron transfer, whereas the active site of the MoFe protein contains the polynuclear FeMo cofactor, a species composed of seven iron atoms, one molybdenum atom, nine sulfur atoms, an interstitial light atom, and one homocitrate molecule. This Perspective provides an overview of biological nitrogen fixation and introduces three contributions to this special feature that address central aspects of the mechanism and assembly of nitrogenase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
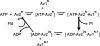
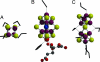
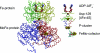
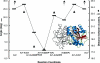
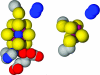
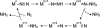
 Mo and Fe, n = +3 and +1, respectively.
Mo and Fe, n = +3 and +1, respectively.References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

