Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif
- PMID: 17090219
- PMCID: PMC1634882
- DOI: 10.1371/journal.pbio.0040377
Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif
Abstract
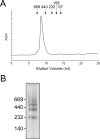
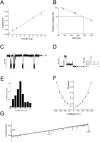
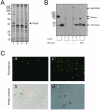
Integral beta-barrel proteins are found in the outer membranes of Gram-negative bacteria, mitochondria, and chloroplasts. The assembly of these proteins requires a proteinaceous apparatus of which Omp85 is an evolutionary conserved central component. To study its molecular mechanism, we have produced Omp85 from Escherichia coli in inclusion bodies and refolded it in vitro. The interaction of Omp85 with its substrate proteins was studied in lipid-bilayer experiments, where it formed channels. The properties of these channels were affected upon addition of unfolded outer-membrane proteins (OMPs) or synthetic peptides corresponding to their C-terminal signature sequences. The interaction exhibited species specificity, explaining the inefficient assembly of OMPs from Neisseria in E. coli. Accordingly, the in vivo assembly of the neisserial porin PorA into the E. coli outer membrane was accomplished after adapting its signature sequence. These results demonstrate that the Omp85 assembly machinery recognizes OMPs by virtue of their C-terminal signature sequence.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures



Comment in
-
Picking the right parts at the beta barrel factory.PLoS Biol. 2006 Nov;4(11):e399. doi: 10.1371/journal.pbio.0040399. Epub 2006 Nov 7. PLoS Biol. 2006. PMID: 20076495 Free PMC article. No abstract available.
References
-
- Koebnik R, Locher KP, Van Gelder P. Structure and function of bacterial outer membrane proteins: Barrels in a nutshell. Mol Microbiol. 2000;37:239–253. - PubMed
-
- Osborne AR, Rapoport TA, Van den Berg B. Protein translocation by the Sec61/SecY channel. Annu Rev Cell Dev Biol. 2005;21:529–550. - PubMed
-
- Harms N, Koningstein G, Dontje W, Müller M, Oudega B, et al. The early interaction of the outer membrane protein PhoE with the periplasmic chaperone Skp occurs at the cytoplasmic membrane. J Biol Chem. 2001;276:18804–18811. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

