Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target
- PMID: 17090670
- PMCID: PMC1635024
- DOI: 10.1073/pnas.0608396103
Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target
Abstract
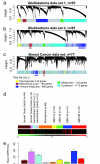
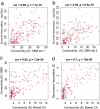
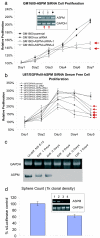
Glioblastoma is the most common primary malignant brain tumor of adults and one of the most lethal of all cancers. Patients with this disease have a median survival of 15 months from the time of diagnosis despite surgery, radiation, and chemotherapy. New treatment approaches are needed. Recent works suggest that glioblastoma patients may benefit from molecularly targeted therapies. Here, we address the compelling need for identification of new molecular targets. Leveraging global gene expression data from two independent sets of clinical tumor samples (n = 55 and n = 65), we identify a gene coexpression module in glioblastoma that is also present in breast cancer and significantly overlaps with the "metasignature" for undifferentiated cancer. Studies in an isogenic model system demonstrate that this module is downstream of the mutant epidermal growth factor receptor, EGFRvIII, and that it can be inhibited by the epidermal growth factor receptor tyrosine kinase inhibitor Erlotinib. We identify ASPM (abnormal spindle-like microcephaly associated) as a key gene within this module and demonstrate its overexpression in glioblastoma relative to normal brain (or body tissues). Finally, we show that ASPM inhibition by siRNA-mediated knockdown inhibits tumor cell proliferation and neural stem cell proliferation, supporting ASPM as a potential molecular target in glioblastoma. Our weighted gene coexpression network analysis provides a blueprint for leveraging genomic data to identify key control networks and molecular targets for glioblastoma, and the principle eluted from our work can be applied to other cancers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Sawyers C. Nature. 2004;432:294–297. - PubMed
-
- Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute DJ, et al. N Engl J Med. 2005;353:2012–2024. - PubMed
-
- Liu ET, Karuturi KR. N Engl J Med. 2004;350:1595–1597. - PubMed
-
- Ebert BL, Golub TR. Blood. 2004;104:923–932. - PubMed
-
- Hood L, Heath JR, Phelps ME, Lin B. Science. 2004;306:640–643. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

