Global mapping of c-Myc binding sites and target gene networks in human B cells
- PMID: 17093053
- PMCID: PMC1635161
- DOI: 10.1073/pnas.0604129103
Global mapping of c-Myc binding sites and target gene networks in human B cells
Abstract
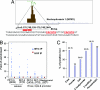
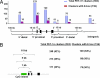
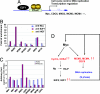
The protooncogene MYC encodes the c-Myc transcription factor that regulates cell growth, cell proliferation, cell cycle, and apoptosis. Although deregulation of MYC contributes to tumorigenesis, it is still unclear what direct Myc-induced transcriptomes promote cell transformation. Here we provide a snapshot of genome-wide, unbiased characterization of direct Myc binding targets in a model of human B lymphoid tumor using ChIP coupled with pair-end ditag sequencing analysis (ChIP-PET). Myc potentially occupies > 4,000 genomic loci with the majority near proximal promoter regions associated frequently with CpG islands. Using gene expression profiles with ChIP-PET, we identified 668 direct Myc-regulated gene targets, including 48 transcription factors, indicating that Myc is a central transcriptional hub in growth and proliferation control. This first global genomic view of Myc binding sites yields insights of transcriptional circuitries and cis regulatory modules involving Myc and provides a substantial framework for our understanding of mechanisms of Myc-induced tumorigenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Adhikary S, Eilers M. Nat Rev Mol Cell Biol. 2005;6:635–645. - PubMed
-
- Grandori C, Cowley SM, James LP, Eisenman RN. Annu Rev Cell Dev Biol. 2000;16:653–699. - PubMed
-
- Blackwood EM, Eisenman RN. Science. 1991;251:1211–1217. - PubMed
-
- Claassen GF, Hann SR. Oncogene. 1999;18:2925–2933. - PubMed
-
- Amati B, Brooks MW, Levy N, Littlewood TD, Evan GI, Land H. Cell. 1993;72:233–245. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

