Cell migration along the lateral cortical stream to the developing basal telencephalic limbic system
- PMID: 17093077
- PMCID: PMC6674782
- DOI: 10.1523/JNEUROSCI.3092-06.2006
Cell migration along the lateral cortical stream to the developing basal telencephalic limbic system
Abstract
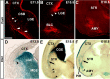
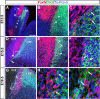
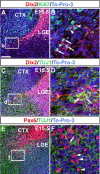
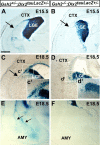
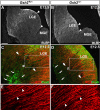
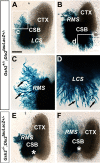
During embryogenesis, the lateral cortical stream (LCS) emerges from the corticostriatal border (CSB), the boundary between the developing cerebral cortex and striatum. The LCS is comprised of a mix of pallial- and subpallial-derived neural progenitor cells that migrate to the developing structures of the basal telencephalon, most notably the piriform cortex and amygdala. Using a combination of in vitro and in vivo approaches, we analyzed the timing, composition, migratory modes, origin, and requirement of the homeodomain-containing transcription factor Gsh2 (genomic screened homeobox 2) in the development of this prominent migratory stream. We reveal that Pax6 (paired box gene 6)-positive pallial-derived and Dlx2 (distal-less homeobox 2)-positive subpallial-derived subpopulations of LCS cells are generated in distinct temporal windows during embryogenesis. Furthermore, our data indicate the CSB border not only is comprised of separate populations of pallial- and subpallial-derived progenitors that contribute to the LCS but also a subpopulation of cells coexpressing Pax6 and Dlx2. Moreover, despite migrating along a route outlined by a cascade of radial glia, the Dlx2-positive population appears to migrate primarily in an apparent chain-like manner, with LCS migratory cells being generated locally at the CSB with little contribution from other subpallial structures such as the medial, lateral, or caudal ganglionic eminences. We further demonstrate that the generation of the LCS is dependent on the homeodomain-containing gene Gsh2, revealing a novel requirement for Gsh2 in telencephalic development.
Figures




References
-
- Alheid GF. Extended amygdala and basal forebrain. Ann NY Acad Sci. 2003;985:185–205. - PubMed
-
- Anthony TE, Klein C, Fishell G, Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. - PubMed
-
- Bayer S, Altman J. Neocortical development. New York: Raven; 1991.
Publication types
MeSH terms
Substances
Grants and funding
- MH068469/MH/NIMH NIH HHS/United States
- MH071679/MH/NIMH NIH HHS/United States
- NS 27054/NS/NINDS NIH HHS/United States
- DA020140/DA/NIDA NIH HHS/United States
- NS032993/NS/NINDS NIH HHS/United States
- T32 HD 007459/HD/NICHD NIH HHS/United States
- R01 NS027054/NS/NINDS NIH HHS/United States
- R01 MH068469/MH/NIMH NIH HHS/United States
- T32 HD007459/HD/NICHD NIH HHS/United States
- R01 NS032993/NS/NINDS NIH HHS/United States
- R01 DA020140/DA/NIDA NIH HHS/United States
- R01 MH071679/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
