Dynamic state of DNA topology is essential for genome condensation in bacteria
- PMID: 17093499
- PMCID: PMC1679767
- DOI: 10.1038/sj.emboj.7601414
Dynamic state of DNA topology is essential for genome condensation in bacteria
Abstract
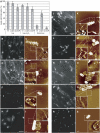
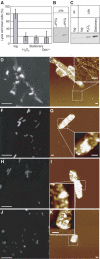
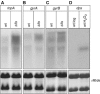
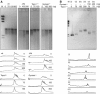
In bacteria, Dps is one of the critical proteins to build up a condensed nucleoid in response to the environmental stresses. In this study, we found that the expression of Dps and the nucleoid condensation was not simply correlated in Escherichia coli, and that Fis, which is an E. coli (gamma-Proteobacteria)-specific nucleoid protein, interfered with the Dps-dependent nucleoid condensation. Atomic force microscopy and Northern blot analyses indicated that the inhibitory effect of Fis was due to the repression of the expression of Topoismerase I (Topo I) and DNA gyrase. In the Deltafis strain, both topA and gyrA/B genes were found to be upregulated. Overexpression of Topo I and DNA gyrase enhanced the nucleoid condensation in the presence of Dps. DNA-topology assays using the cell extract showed that the extracts from the Deltafis and Topo I-/DNA gyrase-overexpressing strains, but not the wild-type extract, shifted the population toward relaxed forms. These results indicate that the topology of DNA is dynamically transmutable and that the topology control is important for Dps-induced nucleoid condensation.
Figures



References
-
- Almiron M, Link AJ, Furlong D, Kolter R (1992) A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev 6: 2646–2654 - PubMed
-
- Altuvia S, Almiron M, Huisman G, Kolter R, Storz G (1994) The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol Microbiol 13: 265–272 - PubMed
-
- Azam TA, Ishihama A (1999) Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J Biol Chem 274: 33105–33113 - PubMed
-
- Azam TA, Hiraga S, Ishihama A (2000) Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells 5: 613–626 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

