The frequency of translational misreading errors in E. coli is largely determined by tRNA competition
- PMID: 17095544
- PMCID: PMC1705757
- DOI: 10.1261/rna.294907
The frequency of translational misreading errors in E. coli is largely determined by tRNA competition
Abstract
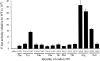
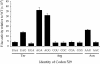
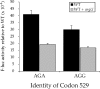
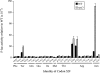
Estimates of missense error rates (misreading) during protein synthesis vary from 10(-3) to 10(-4) per codon. The experiments reporting these rates have measured several distinct errors using several methods and reporter systems. Variation in reported rates may reflect real differences in rates among the errors tested or in sensitivity of the reporter systems. To develop a more accurate understanding of the range of error rates, we developed a system to quantify the frequency of every possible misreading error at a defined codon in Escherichia coli. This system uses an essential lysine in the active site of firefly luciferase. Mutations in Lys529 result in up to a 1600-fold reduction in activity, but the phenotype varies with amino acid. We hypothesized that residual activity of some of the mutant genes might result from misreading of the mutant codons by tRNA(Lys) (UUUU), the cognate tRNA for the lysine codons, AAA and AAG. Our data validate this hypothesis and reveal details about relative missense error rates of near-cognate codons. The error rates in E. coli do, in fact, vary widely. One source of variation is the effect of competition by cognate tRNAs for the mutant codons; higher error frequencies result from lower competition from low-abundance tRNAs. We also used the system to study the effect of ribosomal protein mutations known to affect error rates and the effect of error-inducing antibiotics, finding that they affect misreading on only a subset of near-cognate codons and that their effect may be less general than previously thought.
Figures


References
-
- Amann, E., Ochs, B., Abel, K.J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli . Gene. 1988;69:301–315. - PubMed
-
- Andersson, D.I., Bohman, K., Isaksson, L.A., Kurland, C.G. Translation rates and misreading characteristics of rpsD mutants in Escherichia coli . Mol. Gen. Genet. 1982;187:467–472. - PubMed
-
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., Struhl, K. Current protocols in molecular biology. John Wiley & Sons; New York: 1997.
-
- Branchini, B.R., Murtiashaw, M.H., Magyar, R.A., Anderson, S.M. The role of lysine 529, a conserved residue of the acyl-adenylate-forming enzyme superfamily, in firefly luciferase. Biochemistry. 2000;39:5433–5440. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
