Human-specific insertions and deletions inferred from mammalian genome sequences
- PMID: 17095709
- PMCID: PMC1716262
- DOI: 10.1101/gr.5429606
Human-specific insertions and deletions inferred from mammalian genome sequences
Abstract
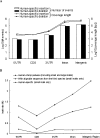
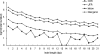
It has been suggested that insertions and deletions (indels) have contributed to the sequence divergence between the human and chimpanzee genomes more than do nucleotide changes (3% vs. 1.2%). However, although there have been studies of large indels between the two genomes, no systematic analysis of small indels (i.e., indels </= 100 bp) has been published. In this study, we first estimated that the false-positive rate of small indels inferred from human-chimpanzee pairwise sequence alignments is quite high, suggesting that the chimpanzee genome draft is not sufficiently accurate for our purpose. We have therefore inferred only human-specific indels using multiple sequence alignments of mammalian genomes. We identified >840,000 "small" indels, which affect >7000 UCSC-annotated human genes (>11,000 transcripts). These indels, however, amount to only approximately 0.21% sequence change in the human lineage for the regions compared, whereas in pseudogenes indels contribute to a sequence divergence of 1.40%, suggesting that most of the indels that occurred in genic regions have been eliminated. Functional analysis reveals that the genes whose coding exons have been affected by human-specific indels are enriched in transcription and translation regulatory activities but are underrepresented in catalytic and transporter activities, cellular and physiological processes, and extracellular region/matrix. This functional bias suggests that human-specific indels might have contributed to human unique traits by causing changes at the RNA and protein level.
Figures



References
-
- The Chimpanzee Genome Sequencing Analysis Consortium, Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. - PubMed
-
- Clark A.G., Glanowski S., Nielsen R., Thomas P.D., Kejariwal A., Todd M.A., Tanenbaum D.M., Civello D., Lu F., Murphy B., Glanowski S., Nielsen R., Thomas P.D., Kejariwal A., Todd M.A., Tanenbaum D.M., Civello D., Lu F., Murphy B., Nielsen R., Thomas P.D., Kejariwal A., Todd M.A., Tanenbaum D.M., Civello D., Lu F., Murphy B., Thomas P.D., Kejariwal A., Todd M.A., Tanenbaum D.M., Civello D., Lu F., Murphy B., Kejariwal A., Todd M.A., Tanenbaum D.M., Civello D., Lu F., Murphy B., Todd M.A., Tanenbaum D.M., Civello D., Lu F., Murphy B., Tanenbaum D.M., Civello D., Lu F., Murphy B., Civello D., Lu F., Murphy B., Lu F., Murphy B., Murphy B., et al. Inferring nonneutral evolution from human–chimp–mouse orthologous gene trios. Science. 2003;302:1960–1963. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
