Conformation of the HIV-1 Gag protein in solution
- PMID: 17097677
- PMCID: PMC1866279
- DOI: 10.1016/j.jmb.2006.10.073
Conformation of the HIV-1 Gag protein in solution
Abstract
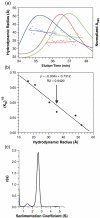
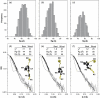
A single multi-domain viral protein, termed Gag, is sufficient for assembly of retrovirus-like particles in mammalian cells. We have purified the human immunodeficiency virus type 1 (HIV-1) Gag protein (lacking myristate at its N terminus and the p6 domain at its C terminus) from bacteria. This protein is capable of assembly into virus-like particles in a defined in vitro system. We have reported that it is in monomer-dimer equilibrium in solution, and have described a mutant Gag protein that remains monomeric at high concentrations in solution. We report that the mutant protein retains several properties of wild-type Gag. This mutant enabled us to analyze solutions of monomeric protein. Hydrodynamic studies on the mutant protein showed that it is highly asymmetric, with a frictional ratio of 1.66. Small-angle neutron scattering (SANS) experiments confirmed its asymmetry and yielded an R(g) value of 34 A. Atomic-level structures of individual domains within Gag have previously been determined, but these domains are connected in Gag by flexible linkers. We constructed a series of models of the mutant Gag protein based on these domain structures, and tested each model computationally for its agreement with the experimental hydrodynamic and SANS data. The only models consistent with the data were those in which Gag was folded over, with its N-terminal matrix domain near its C-terminal nucleocapsid domain in three-dimensional space. Since Gag is a rod-shaped molecule in the assembled immature virion, these findings imply that Gag undergoes a major conformational change upon virus assembly.
Figures



References
-
- Swanstrom R, Wills JW. Synthesis, assembly, and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retro-viruses. Cold Spring Harbor Laboratory Press; Plainview, NY: 1997. pp. 263–334. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

