The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis
- PMID: 17098808
- PMCID: PMC1693934
- DOI: 10.1105/tpc.106.045617
The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis
Erratum in
-
Correction to: The Balance between the MIR164A and CUC2 Genes Controls Leaf Margin Serration in Arabidopsis.Plant Cell. 2025 Jun 4;37(6):koaf123. doi: 10.1093/plcell/koaf123. Plant Cell. 2025. PMID: 40476682 Free PMC article. No abstract available.
Abstract
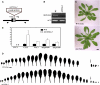
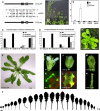
CUP-SHAPED COTYLEDON1 (CUC1), CUC2, and CUC3 define the boundary domain around organs in the Arabidopsis thaliana meristem. CUC1 and CUC2 transcripts are targeted by a microRNA (miRNA), miR164, encoded by MIR164A, B, and C. We show that each MIR164 is transcribed to generate a large population of primary miRNAs of variable size with a locally conserved secondary structure around the pre-miRNA. We identified mutations in the MIR164A gene that deepen serration of the leaf margin. By contrast, leaves of plants overexpressing miR164 have smooth margins. Enhanced leaf serration was observed following the expression of an miR164-resistant CUC2 but not of an miR164-resistant CUC1. Furthermore, CUC2 inactivation abolished serration in mir164a mutants and the wild type, whereas CUC1 inactivation did not. Thus, CUC2 specifically controls leaf margin development. CUC2 and MIR164A are transcribed in overlapping domains at the margins of young leaf primordia, with transcription gradually restricted to the sinus, where the leaf margins become serrated. We suggest that leaf margin development is controlled by a two-step process in Arabidopsis. The pattern of serration is determined first, independently of CUC2 and miR164. The balance between coexpressed CUC2 and MIR164A then determines the extent of serration.
Figures




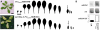


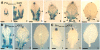

References
-
- Aida, M., Ishida, T., and Tasaka, M. (1999). Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126 1563–1570. - PubMed
-
- Aloni, R., Schwalm, K., Langhans, M., and Ullrich, C.I. (2003). Gradual shifts in sites of free-auxin production during leaf-primordium development and their role in vascular differentiation and leaf morphogenesis in Arabidopsis. Planta 216 841–853. - PubMed
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

